7KPH
 
 | | SARS-CoV-2 Main Protease in mature form | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-11-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7KVR
 
 | | SARS-CoV-2 Main protease immature form - FMAX Library E09 fragment | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7KFI
 
 | | SARS-CoV-2 Main protease immature form - apo structure | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-10-14 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7KVL
 
 | | SARS-CoV-2 Main protease immature form - FMAX Library E01 fragment | | Descriptor: | 2-chloropyridine-4-carboxamide, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7LFP
 
 | | SARS-CoV-2 Main protease immature form - F2X Entry Library G05 fragment | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7LDX
 
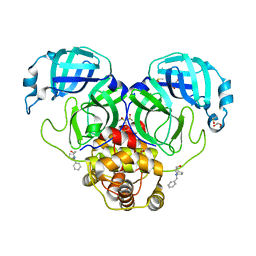 | | SARS-CoV-2 Main protease immature form - F2X Entry Library E06 fragment | | Descriptor: | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7LFE
 
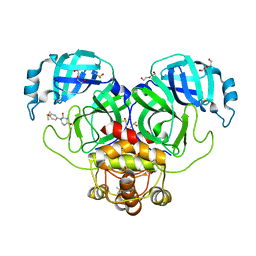 | | SARS-CoV-2 Main protease immature form - F2X Entry Library E03 fragment | | Descriptor: | (2R,4R)-1-phenylhexahydropyrimidine-2,4-diol, 3C-like proteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-01-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7MBG
 
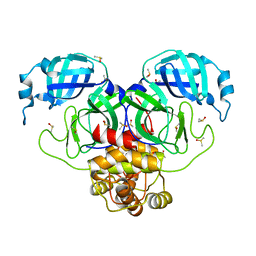 | | SARS-CoV-2 Main protease in orthorhombic space group | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Douangamath, A, von Delft, F, Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KF4
 
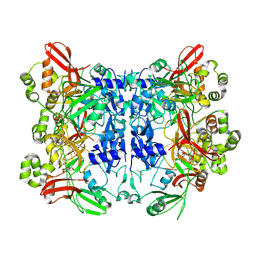 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KEH
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KEG
 
 | | Crystal structure from SARS-COV2 NendoU NSP15 | | Descriptor: | PHOSPHATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
1II2
 
 | | Crystal Structure of Phosphoenolpyruvate Carboxykinase (PEPCK) from Trypanosoma cruzi | | Descriptor: | PHOSPHOENOLPYRUVATE CARBOXYKINASE, SULFATE ION | | Authors: | Trapani, S, Linss, J, Goldenberg, S, Fischer, H, Craievich, A.F, Oliva, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the dimeric phosphoenolpyruvate carboxykinase (PEPCK) from Trypanosoma cruzi at 2 A resolution.
J.Mol.Biol., 313, 2001
|
|
1ML3
 
 | | Evidences for a flip-flop catalytic mechanism of Trypanosoma cruzi glyceraldehyde-3-phosphate dehydrogenase, from its crystal structure in complex with reacted irreversible inhibitor 2-(2-phosphono-ethyl)-acrylic acid 4-nitro-phenyl ester | | Descriptor: | (3-FORMYL-BUT-3-ENYL)-PHOSPHONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Castilho, M.S, Pavao, F, Oliva, G. | | Deposit date: | 2002-08-29 | | Release date: | 2003-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for the Two Phosphate Binding Sites of an Analogue of the Thioacyl Intermediate for the Trypanosoma cruzi Glyceraldehyde-3-phosphate Dehydrogenase-Catalyzed Reaction, from Its Crystal Structure.
Biochemistry, 42, 2003
|
|
8DZ0
 
 | | Crystal Structure of SARS-CoV-2 Main protease in complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZA
 
 | | Crystal Structure of SARS-CoV-2 Main protease A193T mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ1
 
 | | Crystal Structure of SARS-CoV-2 Main protease mutant M49I in complex with Ensitrelvir | | Descriptor: | 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ6
 
 | | Crystal Structure of SARS-CoV-2 Main protease mutant Q189K in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.366 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8DZ2
 
 | | Crystal Structure of SARS-CoV-2 Main protease in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8EY2
 
 | | Cryo-EM structure of SARS-CoV-2 Main protease C145S in complex with N-terminal peptide | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Song, Y, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An in-solution snapshot of SARS-COV-2 main protease maturation process and inhibition.
Nat Commun, 14, 2023
|
|
5U04
 
 | | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase | | Descriptor: | PHOSPHATE ION, ZINC ION, Zika Virus NS5 RdRp | | Authors: | Godoy, A.S, Lima, G.M.A, Oliveira, K.I.Z, Torres, N.U, Maluf, F.V, Guido, R.V.C, Oliva, G. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase.
Nat Commun, 8, 2017
|
|
8DZ9
 
 | | Crystal Structure of SARS-CoV-2 Main protease G143S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E1Y
 
 | | Crystal Structure of SARS-CoV-2 Main protease A193S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
5RHM
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z1454310449 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, N-[(2-fluorophenyl)methyl]-1H-pyrazol-4-amine, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RHX
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z1324080698 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-fluoro-5-methylbenzene-1-sulfonamide, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
