2K21
 
 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2LLE
 
 | | Computational design of an eight-stranded (beta/alpha)-barrel from fragments of different folds | | Descriptor: | Chemotaxis protein CheY, Imidazole glycerol phosphate synthase subunit HisF chimera | | Authors: | Coles, M, Truffault, V, Eisenbeis, S, Proffitt, W, Meiler, J, Hocker, B. | | Deposit date: | 2011-11-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Potential of fragment recombination for rational design of proteins.
J.Am.Chem.Soc., 134, 2012
|
|
2K39
 
 | | Recognition dynamics up to microseconds revealed from RDC derived ubiquitin ensemble in solution | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Fares, C, Schroder, G, Walter, K, Becker, S, Meiler, J, Grubmuller, H, Griesinger, C, de Groot, B.L. | | Deposit date: | 2008-04-25 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution.
Science, 320, 2008
|
|
3MWL
 
 | | Q28E mutant of HERA N-terminal RecA-like domain in complex with 8-OXOADENOSINE | | Descriptor: | 6-azanyl-9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-7H-purin-8-one, Heat resistant RNA dependent ATPase, SULFATE ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3MWJ
 
 | |
3MWK
 
 | | Q28E mutant of HERA N-terminal RecA-like domain, complex with 8-oxo-AMP | | Descriptor: | Heat resistant RNA dependent ATPase, SULFATE ION, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3NBF
 
 | | Q28E mutant of hera helicase N-terminal domain bound to 8-oxo-ADP | | Descriptor: | Heat resistant RNA dependent ATPase, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl dihydrogen phosphate, [(2R,3S,4R,5R)-5-(6-azanyl-8-oxo-7H-purin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2010-06-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changing nucleotide specificity of the DEAD-box helicase Hera abrogates communication between the Q-motif and the P-loop.
Biol.Chem., 392, 2011
|
|
3OBR
 
 | |
3OBT
 
 | |
3NEJ
 
 | |
5O30
 
 | |
6DCW
 
 | |
4ZV6
 
 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
6S4A
 
 | | Structure of human MTHFD2 in complex with TH9028 | | Descriptor: | (2~{S})-2-[[5-[[2,4-bis(azanyl)-6-oxidanylidene-5~{H}-pyrimidin-5-yl]carbamoylamino]pyridin-2-yl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4F
 
 | | Structure of human MTHFD2 in complex with TH9619 | | Descriptor: | (E,4S)-4-[[5-[2-[2,6-bis(azanyl)-4-oxidanylidene-1H-pyrimidin-5-yl]ethanoylamino]-3-fluoranyl-pyridin-2-yl]carbonylamino]pent-2-enedioic acid, ADENOSINE-5'-DIPHOSPHATE, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Scaletti, E.R, Gustafsson, R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4E
 
 | | Structure of human MTHFD2 in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
3MFA
 
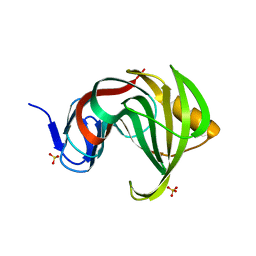 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
7ZGM
 
 | | Plant N-glycan specific alpha-1,3-mannosidase | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 92 protein, POTASSIUM ION, ... | | Authors: | Basle, A, Crouch, L, Bolam, D. | | Deposit date: | 2022-04-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Plant N -glycan breakdown by human gut Bacteroides.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZGN
 
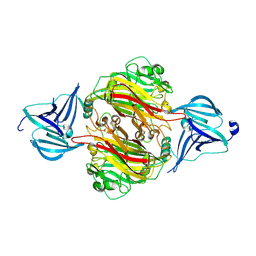 | |
4ZWQ
 
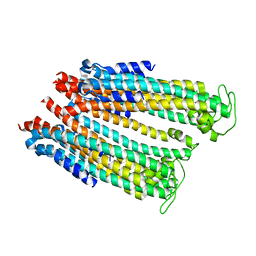 | |
4ZWR
 
 | |
4ZWS
 
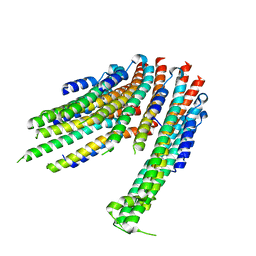 | |
4ZWT
 
 | |
6E1F
 
 | |
7SC0
 
 | |
