8K1D
 
 | | SID1 transmembrane family member 1 | | Descriptor: | SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
7QXX
 
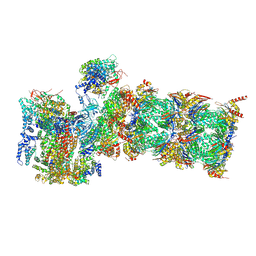 | | Proteasome-ZFAND5 Complex Z+E state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QY7
 
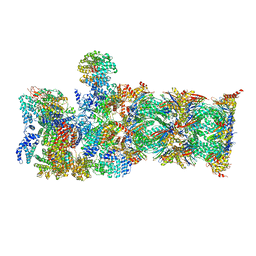 | | Proteasome-ZFAND5 Complex Z-A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXN
 
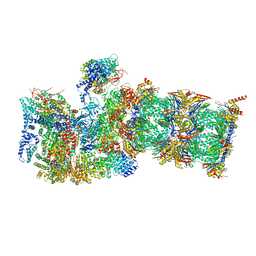 | | Proteasome-ZFAND5 Complex Z+A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXP
 
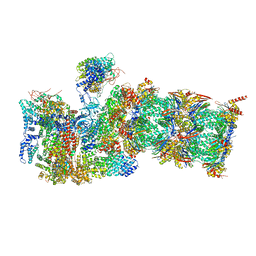 | | Proteasome-ZFAND5 Complex Z+B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QYB
 
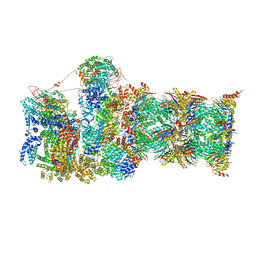 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXU
 
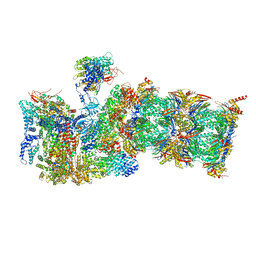 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QXW
 
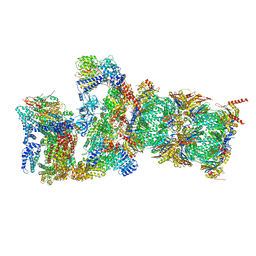 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
7QYA
 
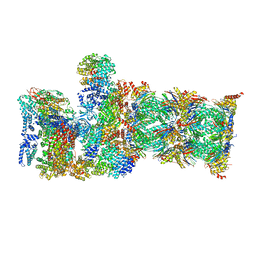 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanism of 26S proteasome activation by the 19S-interacting protein ZFAND5
To Be Published
|
|
4TYX
 
 | | Structure of aquoferric sperm whale myoglobin L29H/F33Y/F43H/S92A mutant | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhagi-Damodaran, A, Petrik, I.D, Robinson, H, Lu, Y. | | Deposit date: | 2014-07-09 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Systematic tuning of heme redox potentials and its effects on O2 reduction rates in a designed oxidase in myoglobin.
J.Am.Chem.Soc., 136, 2014
|
|
1CC3
 
 | | PURPLE CUA CENTER | | Descriptor: | COPPER (II) ION, PROTEIN (CUA AZURIN) | | Authors: | Robinson, H, Ang, M.C, Gao, Y.-G, Hay, M.T, Lu, Y, Wang, A.H.-J. | | Deposit date: | 1999-03-03 | | Release date: | 1999-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of electron transfer modulation in the purple CuA center.
Biochemistry, 38, 1999
|
|
4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
1LUF
 
 | | Crystal Structure of the MuSK Tyrosine Kinase: Insights into Receptor Autoregulation | | Descriptor: | muscle-specific tyrosine kinase receptor musk | | Authors: | Till, J.H, Becerra, M, Watty, A, Lu, Y, Ma, Y, Neubert, T.A, Burden, S.J, Hubbard, S.R. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation.
Structure, 10, 2002
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | Descriptor: | A-BETA AMYLOID | | Authors: | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
7TNC
 
 | | M13F/G116F Pseudomonas aeruginosa azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, CHLORIDE ION, ... | | Authors: | Liu, Y, Lu, Y. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for the Effects of Phenylalanine on Tuning the Reduction Potential of Type 1 Copper in Azurin.
Inorg.Chem., 62, 2023
|
|
7U2F
 
 | | G116F Pseudomonas aeruginosa azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Liu, Y, Lu, Y. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Effects of Phenylalanine on Tuning the Reduction Potential of Type 1 Copper in Azurin.
Inorg.Chem., 62, 2023
|
|
8U8Q
 
 | | V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8P
 
 | | S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8R
 
 | | Y229F/V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8S
 
 | | Y229F/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8T
 
 | | Y229F/V290N Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4WKX
 
 | |
8Y7Y
 
 | | Local structure of HCoV-HKU1A spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y, Liu, X, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
4HK4
 
 | | Crystal structure of apo Tyrosine-tRNA ligase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal structure of apo Tyrosine-tRNA ligase mutant protein
To be Published
|
|
