8G6V
 
 | |
8G8Y
 
 | |
8GCN
 
 | |
4UAF
 
 | |
4UAM
 
 | | 1.8 Angstrom crystal structure of IMP-1 metallo-beta-lactamase with a mixed iron-zinc center in the active site | | Descriptor: | CITRATE ANION, FE (III) ION, IMP-1 metallo-beta-lactamase, ... | | Authors: | Carruthers, T.J, Carr, P.D, Jackson, C.J, Otting, G. | | Deposit date: | 2014-08-11 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Iron(III) Located in the Dinuclear Metallo-beta-Lactamase IMP-1 by Pseudocontact Shifts.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UAD
 
 | |
4UAE
 
 | |
5HIE
 
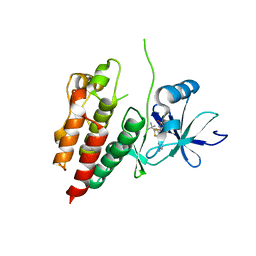 | | BRAF Kinase domain b3aC loop deletion mutant in complex with dabrafenib | | Descriptor: | Dabrafenib, Serine/threonine-protein kinase B-raf | | Authors: | Whalen, D.M, Foster, S.A, Ozen, A, Wongchenko, M, Yin, J, Schaefer, G, Mayfield, J, Chmielecki, J, Stephens, P, Albacker, L, Yan, Y, Song, K, Hatzivassiliou, G, Eigenbrot, C, Yu, C, Shaw, A.S, Manning, G, Skelton, N.J, Hymowitz, S.G, Malek, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
4XZR
 
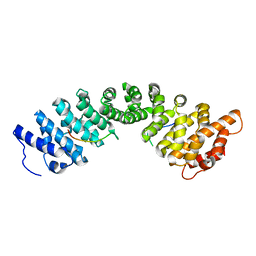 | |
4YI0
 
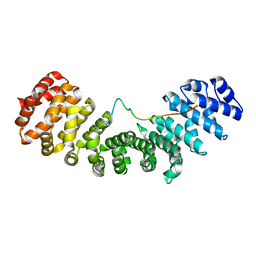 | | Structure of mouse importin a1 bound to Pom121NLS | | Descriptor: | Importin subunit alpha-1, Nuclear envelope pore membrane protein POM 121 | | Authors: | Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2015-02-27 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Mammalian Pom121 and yeast Heh2 share IBB-like NLSs that support targeting to the inner nuclear membrane
To Be Published
|
|
4ZKU
 
 | | P22 Tail Needle Gp26 crystallized at pH 10.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tail needle protein gp26 | | Authors: | Sankhala, R.S, Cingolani, G. | | Deposit date: | 2015-04-30 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Plasticity of the Protein Plug That Traps Newly Packaged Genomes in Podoviridae Virions.
J.Biol.Chem., 291, 2016
|
|
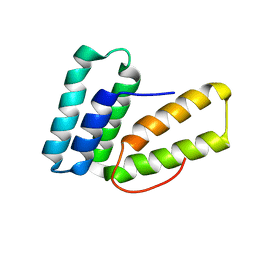
6O6I
 
 | |
8G83
 
 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | Descriptor: | NAD(+) hydrolase AbTIR | | Authors: | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | Deposit date: | 2023-02-17 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
5HUY
 
 | |
5HUW
 
 | |
8EON
 
 | | Pseudomonas phage E217 baseplate complex | | Descriptor: | Baseplate component gp33, Baseplate component gp34, Baseplate component gp36, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
8ENV
 
 | | In situ cryo-EM structure of Pseudomonas phage E217 tail baseplate in C6 map | | Descriptor: | Baseplate_J domain-containing protein gp44, Ripcord gp36, Sheath initiator gp34, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
5MW2
 
 | | CRYSTAL STRUCTURE OF BCL-6 BTB-domain with BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
8FUV
 
 | |
8FVG
 
 | |
8FRS
 
 | |
8FVH
 
 | | Pseudomonas phage E217 neck (portal, head-to-tail connector, collar and gateway proteins) | | Descriptor: | E217 collar protein gp28, E217 gateway protein gp29, E217 head-to-tail connector protein gp27, ... | | Authors: | Li, F, Cingolani, G, Hou, C. | | Deposit date: | 2023-01-18 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217.
Nat Commun, 14, 2023
|
|
1AHD
 
 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF AN ANTENNAPEDIA HOMEODOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), Homeotic protein antennapedia | | Authors: | Billeter, M, Qian, Y.Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex.
J.Mol.Biol., 234, 1993
|
|
5MWD
 
 | | Crystal structure of the BCL6 BTB-domain with compound 2 | | Descriptor: | 5-[[5-chloranyl-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
5JJ1
 
 | |
