4JUY
 
 | | Crystal structure of the PUB domain of E3 ubiquitin ligase RNF31 | | Descriptor: | E3 ubiquitin-protein ligase RNF31, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Hu, J, Li, Y, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-25 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the PUB domain of E3 ubiquitin ligase RNF31
To be Published
|
|
4FO9
 
 | | Crystal structure of the E3 SUMO Ligase PIAS2 | | Descriptor: | E3 SUMO-protein ligase PIAS2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Hu, J, Dobrovetsky, E, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the E3 SUMO Ligase PIAS2
to be published
|
|
7X4B
 
 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
5YDT
 
 | | Remodeled Utp30 in 90S pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 5' ETS RNA, Ribosome biogenesis protein UTP30, Saccharomyces cerevisiae strain ALI 308 18S ribosomal RNA gene, ... | | Authors: | Ye, K, Zhu, X, Hu, J. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
6CN3
 
 | |
3NYJ
 
 | | Crystal Structure Analysis of APP E2 domain | | Descriptor: | Amyloid beta A4 protein, OSMIUM ION | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
4MQS
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, Nanobody 9-8 | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
5CB2
 
 | | the structure of candida albicans Sey1p in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Protein SEY1 | | Authors: | Yan, L, Sun, S, Wang, W, Shi, J, Hu, X, Wang, S, Rao, Z, Hu, J, Lou, Z. | | Deposit date: | 2015-06-30 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion
J.Cell Biol., 210, 2015
|
|
4MQT
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo and allosteric modulator LY2119620 | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, ... | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
6PGI
 
 | | Asymmetric functions of a binuclear metal cluster within the transport pathway of the ZIP transition metal transporters | | Descriptor: | BbZIP, CADMIUM ION | | Authors: | Zhang, T, Sui, D, Zhang, C, Logan, T, Hu, J. | | Deposit date: | 2019-06-24 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetric functions of a binuclear metal center within the transport pathway of a human zinc transporter ZIP4.
Faseb J., 34, 2020
|
|
6V0A
 
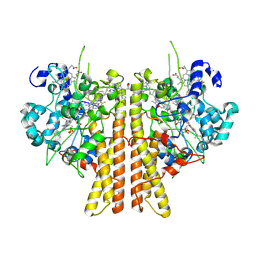 | | Crystal structure of cytochrome c nitrite reductase from the bacterium Geobacter lovleyi with bound sulfate | | Descriptor: | HEME C, Nitrite reductase (cytochrome; ammonia-forming), SULFATE ION | | Authors: | Satyanarayana, L, Campecino, J, Hegg, L.H, Hu, J. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cytochromecnitrite reductase from the bacteriumGeobacter lovleyirepresents a new NrfA subclass.
J.Biol.Chem., 295, 2020
|
|
7S91
 
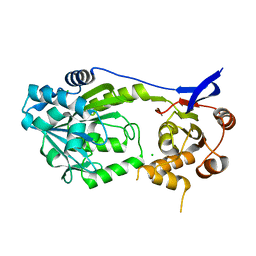 | |
8SOQ
 
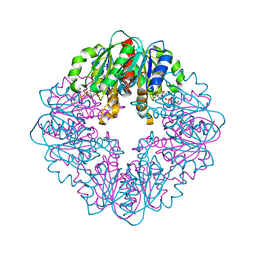 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with authentic substrate NaAD | | Descriptor: | MAGNESIUM ION, NICOTINIC ACID ADENINE DINUCLEOTIDE, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-04-29 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the LarB-Substrate Complex and Identification of a Reaction Intermediate during Nickel-Pincer Nucleotide Cofactor Biosynthesis.
Biochemistry, 62, 2023
|
|
5C7M
 
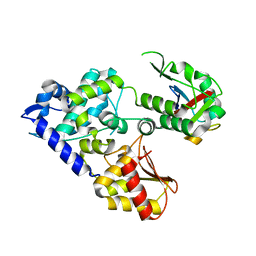 | | CRYSTAL STRUCTURE OF E3 LIGASE ITCH WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Wernimont, A, Zhang, W, Sidhu, S, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
5GNS
 
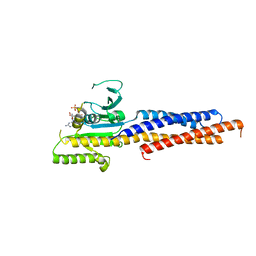 | | Structures of human Mitofusin 1 provide insight into mitochondrial tethering | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mitofusin-1 | | Authors: | Qi, Y, Yan, L, Yu, C, Guo, X, Zhou, X, Hu, X, Huang, X, Rao, Z, Lou, Z, Hu, J. | | Deposit date: | 2016-07-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structures of human Mitofusin 1 provide insight into mitochondrial tethering
To Be Published
|
|
5C7J
 
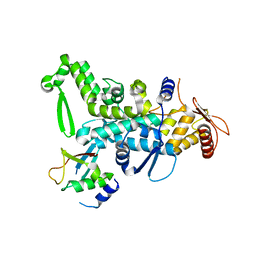 | | CRYSTAL STRUCTURE OF NEDD4 WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
1XZ0
 
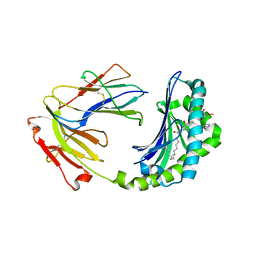 | | Crystal structure of CD1a in complex with a synthetic mycobactin lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(HYDROXY-HEXADECANOYL-AMINO)-2-{[(4S)-2-(2-HYDROXY-PHENYL)-4,5-DIHYDRO-OXAZOLE-4-CARBONYL]-AMINO}-HEXANOIC ACID 2-[(3S)-1-(TERT-BUTYL-DIPHENYL-SILANYLOXY)-2-OXO-AZEPAN-3-YLCARBAMOYL]-(1S)-1-METHYL-ETHYL ESTER, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Crispin, M.D, Bowden, T.A, Young, D.C, Cheng, T.Y, Hu, J, Costello, C.E, Miller, M.J, Moody, D.B, Wilson, I.A. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of Lipopeptide Presentation by CD1a.
Immunity, 22, 2005
|
|
3FP4
 
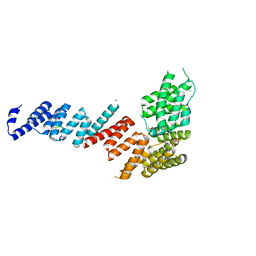 | | Crystal structure of Tom71 complexed with Ssa1 C-terminal fragment | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
6XJI
 
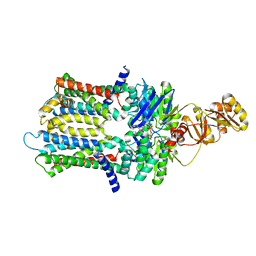 | | PmtCD ABC exporter at C1 symmetry | | Descriptor: | ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zeytuni, N, Strynadka, N.J.C, Hu, J, Worrall, L.J, Chou, H, Yu, Z. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
6OC5
 
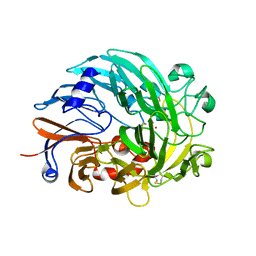 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
6OC6
 
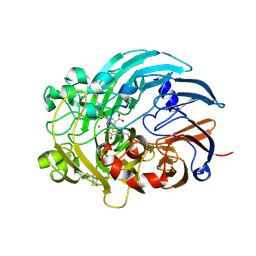 | | Lanthanide-dependent methanol dehydrogenase XoxF from Methylobacterium extorquens, in complex with Lanthanum and Pyrroloquinoline quinone | | Descriptor: | LANTHANUM (III) ION, Lanthanide-dependent methanol dehydrogenase XoxF, PYRROLOQUINOLINE QUINONE | | Authors: | Fellner, M, Good, N.M, Martinez-Gomez, N.C, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function.
J.Biol.Chem., 295, 2020
|
|
2AUG
 
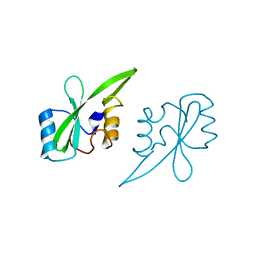 | | Crystal structure of the Grb14 SH2 domain | | Descriptor: | Growth factor receptor-bound protein 14 | | Authors: | Depetris, R.S, Hu, J, Gimpelevich, I, Holt, L.J, Daly, R.J, Hubbard, S.R. | | Deposit date: | 2005-08-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of the insulin receptor by the adaptor protein grb14.
Mol.Cell, 20, 2005
|
|
2AUH
 
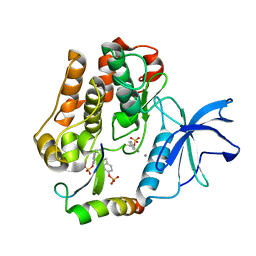 | | Crystal structure of the Grb14 BPS region in complex with the insulin receptor tyrosine kinase | | Descriptor: | CALCIUM ION, Growth factor receptor-bound protein 14, Insulin receptor | | Authors: | Depetris, R.S, Hu, J, Gimpelevich, I, Holt, L.J, Daly, R.J, Hubbard, S.R. | | Deposit date: | 2005-08-27 | | Release date: | 2005-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of the insulin receptor by the adaptor protein grb14.
Mol.Cell, 20, 2005
|
|
6XJH
 
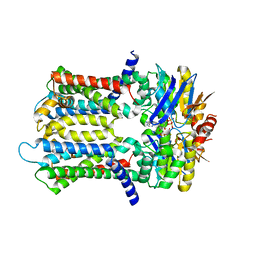 | | PmtCD ABC exporter without the basket domain at C2 symmetry | | Descriptor: | ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Zeytuni, N, Strynadka, N.J.C, Hu, J, Worrall, L.J, Chou, H, Yu, Z. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
3FP2
 
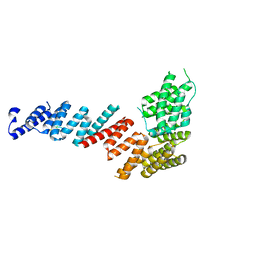 | | Crystal structure of Tom71 complexed with Hsp82 C-terminal fragment | | Descriptor: | ATP-dependent molecular chaperone HSP82, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, J, Qian, X, Hu, J, Sha, B. | | Deposit date: | 2009-01-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading.
J.Biol.Chem., 284, 2009
|
|
