9EOJ
 
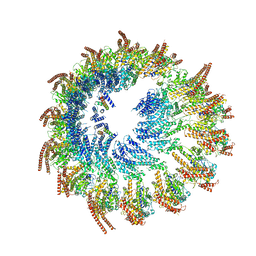 | | Vertebrate microtubule-capping gamma-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component, Gamma-tubulin complex component 3 homolog, Gamma-tubulin complex component 6, ... | | Authors: | Vermeulen, B.J.A, Pfeffer, S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | gamma-TuRC asymmetry induces local protofilament mismatch at the RanGTP-stimulated microtubule minus end.
Embo J., 43, 2024
|
|
9DOJ
 
 | | Influenza A virus polymerase PA subunit endonuclease domain bound to inhibitor Compound 19a | | Descriptor: | (2aS,4aR,10bS,11S,12R)-4a-(2,2-difluoroethyl)-16-hydroxy-2-methyl-2,2a,3,4,4a,5,6,10b-octahydrobenzo[h]pyrido[1',2':1,6][1,2,4]triazino[2,3-a]quinoline-1,15-dione, MALONATE ION, MANGANESE (II) ION, ... | | Authors: | Hollenstein, K, Price, I.R, Eddins, M.J, Su, H.P. | | Deposit date: | 2024-09-19 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Highly Diastereoselective Access to Densely Functionalized Piperidine Cores of Influenza Endonuclease Inhibitors via a Metal-Free S N 1 Approach.
J.Org.Chem., 90, 2025
|
|
9DN4
 
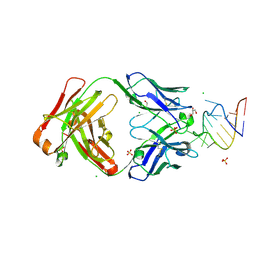 | | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N | | Descriptor: | CHLORIDE ION, FAB BL3-6S97N HEAVY CHAIN, FAB BL3-6S97N LIGHT CHAIN, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Hegde, S, Wang, J. | | Deposit date: | 2024-09-16 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic studies of small molecule ligands selective to RNA single G bulges.
Nucleic Acids Res., 53, 2025
|
|
6BQJ
 
 | |
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
6JYJ
 
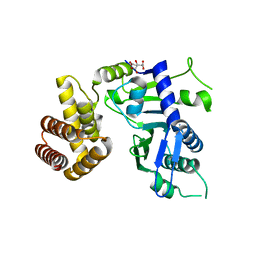 | | Crystal structure of FAM46B (TENT5B) | | Descriptor: | CITRATE ANION, Terminal nucleotidyltransferase 5B | | Authors: | Zhang, H, Hu, J.L, Gao, S. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.693199 Å) | | Cite: | FAM46B is a prokaryotic-like cytoplasmic poly(A) polymerase essential in human embryonic stem cells.
Nucleic Acids Res., 48, 2020
|
|
1D17
 
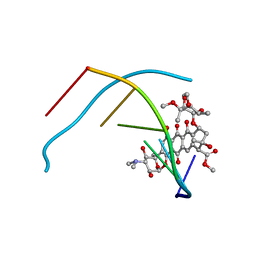 | | DNA-NOGALAMYCIN INTERACTIONS | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*AP*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Egli, M, Williams, L.D, Frederick, C.A, Rich, A. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-nogalamycin interactions.
Biochemistry, 30, 1991
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
131D
 
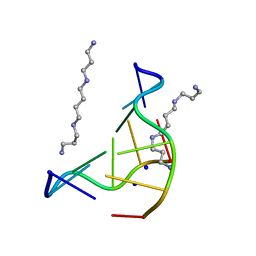 | | THE LOW-TEMPERATURE CRYSTAL STRUCTURE OF THE PURE-SPERMINE FORM OF Z-DNA REVEALS BINDING OF A SPERMINE MOLECULE IN THE MINOR GROOVE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SODIUM ION, SPERMINE | | Authors: | Bancroft, D, Williams, L.D, Rich, A, Egli, M. | | Deposit date: | 1993-06-18 | | Release date: | 1993-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove.
Biochemistry, 33, 1994
|
|
5WSN
 
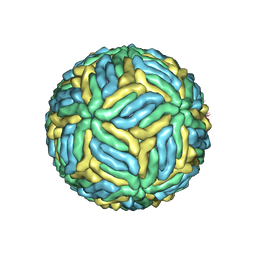 | | Structure of Japanese encephalitis virus | | Descriptor: | E protein, M protein | | Authors: | Wang, X, Zhu, L, Li, S, Yuan, S, Qin, C, Fry, E.E, Stuart, I.D, Rao, Z. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability
Nat Commun, 8, 2017
|
|
4NWK
 
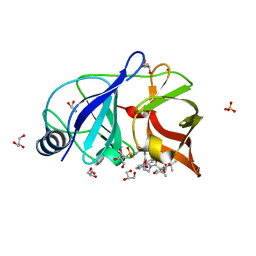 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | Descriptor: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
4NWL
 
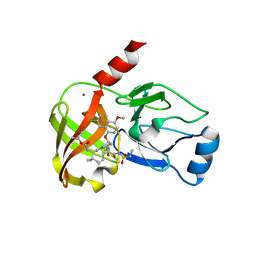 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-650032 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-4-((7-chloro-4-methoxy-1-isoquinolinyl)o xy)-n-((1r,2s)-1-((cyclopropylsulfonyl)carbamoyl)-2-vinylc yclopropyl)-l-prolinamide | | Descriptor: | HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, ZINC ION | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
2F8W
 
 | | Crystal structure of d(CACGTG)2 | | Descriptor: | 1,3-DIAMINOPROPANE, 5'-D(*CP*AP*CP*GP*TP*G)-3', SPERMINE | | Authors: | Narayana, N, Shamala, N, Ganesh, K.N, Viswamitra, M.A. | | Deposit date: | 2005-12-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interaction between the Z-Type DNA Duplex and 1,3-Propanediamine: Crystal Structure of d(CACGTG)2 at 1.2 A Resolution
Biochemistry, 45, 2006
|
|
6LOI
 
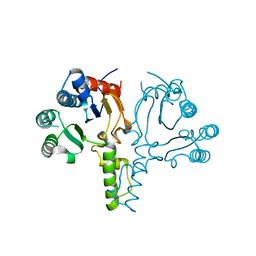 | |
6LQD
 
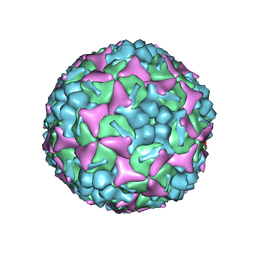 | | Structure of Enterovirus 71 in complex with NLD-22 | | Descriptor: | 1-(2-azanylpyridin-4-yl)-3-[5-[4-(5-methyl-1,2,4-oxadiazol-3-yl)phenoxy]pentyl]imidazolidin-2-one, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Zhang, M, Sun, Y, Wang, X, Guo, Y, Rao, Z. | | Deposit date: | 2020-01-13 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.264 Å) | | Cite: | Design, Synthesis, and Evaluation of Novel Enterovirus 71 Inhibitors as Therapeutic Drug Leads for the Treatment of Human Hand, Foot, and Mouth Disease.
J.Med.Chem., 63, 2020
|
|
8JB4
 
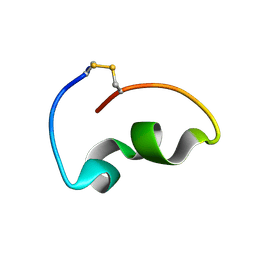 | | lipopolysaccharide-binding domain-LBDB | | Descriptor: | Antilipopolysaccharide factor D | | Authors: | Huang, J, Qin, Z. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-13 | | Last modified: | 2025-03-19 | | Method: | SOLUTION NMR | | Cite: | Machine learning and genetic algorithm-guided directed evolution for the development of antimicrobial peptides.
J Adv Res, 68, 2025
|
|
9JX5
 
 | | Solution NMR structure of the 1:1 complex of NOP56 intronic G-quadruplex bound with pyridostatin | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-azanylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, DNA (5'-D(*GP*GP*GP*CP*CP*TP*GP*GP*GP*CP*CP*TP*GP*GP*GP*CP*CP*TP*GP*GP*G)-3') | | Authors: | Yan, Z, Wan, L, Guo, P, Han, D. | | Deposit date: | 2024-10-11 | | Release date: | 2025-02-05 | | Last modified: | 2025-05-07 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into an Antiparallel Chair-Type G-Quadruplex From the Intron of NOP56 Oncogene.
Adv Sci, 12, 2025
|
|
9FGP
 
 | |
7WEX
 
 | |
5Y9V
 
 | | Crystal structure of diamondback moth ryanodine receptor N-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Ryanodine receptor 1 | | Authors: | Lin, L, Yuchi, Z. | | Deposit date: | 2017-08-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Crystal structure of ryanodine receptor N-terminal domain from Plutella xylostella reveals two potential species-specific insecticide-targeting sites.
Insect Biochem. Mol. Biol., 92, 2017
|
|
5ZUD
 
 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with D6 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
5ZUF
 
 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with A9 | | Descriptor: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | Authors: | Wang, X, Zhu, L, Wang, N. | | Deposit date: | 2018-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
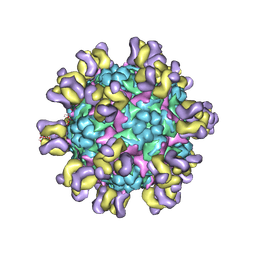
5ZXL
 
 | | Structure of GldA from E.coli | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol dehydrogenase, ... | | Authors: | Zhang, J, Lin, L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structure of glycerol dehydrogenase (GldA) from Escherichia coli.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
6BQK
 
 | |
