6RYO
 
 | | Bacterial membrane enzyme structure by the in meso method at 1.9 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, GLYCEROL, ... | | Authors: | Huang, C.Y, Olatunji, S, Bailey, J, Yu, X, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.924 Å) | | Cite: | Structures of lipoprotein signal peptidase II from Staphylococcus aureus complexed with antibiotics globomycin and myxovirescin.
Nat Commun, 11, 2020
|
|
5N6L
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt C387A mutant from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5N6H
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
5N6M
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5JKI
 
 | | Crystal structure of the first transmembrane PAP2 type phosphatidylglycerolphosphate phosphatase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative lipid phosphate phosphatase YodM, TUNGSTATE(VI)ION, ... | | Authors: | El Ghachi, M, Howe, N, Lampion, A, Delbrassine, F, Vogeley, L, Caffrey, M, Sauvage, E, Auger, R, Guiseppe, A, Roure, S, Perlier, S, Mengin-lecreulx, D, Foglino, M, Touze, T. | | Deposit date: | 2016-04-26 | | Release date: | 2017-02-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure and biochemical characterization of the transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase from Bacillus subtilis.
Cell. Mol. Life Sci., 74, 2017
|
|
6YOE
 
 | | Structure of Lysozyme from SiN IMISX setup collected by still serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
6YOC
 
 | | Structure of Lysozyme from COC IMISX setup collected by still serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
6YOF
 
 | | Structure of PepTSt from COC IMISX setup collected by rotation serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 2-(2-METHOXYETHOXY)ETHANOL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
6YOB
 
 | | Structure of Lysozyme from COC IMISX setup collected by rotation serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
6YOG
 
 | | Structure of PepTSt from COC IMISX setup collected by still serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 2-(2-METHOXYETHOXY)ETHANOL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
6YOD
 
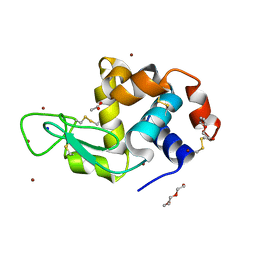 | | Structure of Lysozyme from SiN IMISX setup collected by rotation serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-(2-METHOXYETHOXY)ETHANOL, ACETIC ACID, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
5OON
 
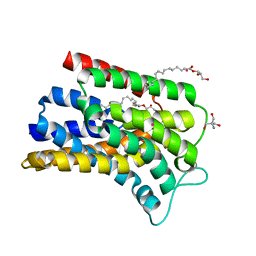 | | Structure of Undecaprenyl-Pyrophosphate Phosphatase, BacA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Wang, M, Howe, N, Ghachi, M.E.I, Weichert, D, Kerff, F, Stansfeld, P, Touze, T, Caffrey, M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis.
Nat Commun, 9, 2018
|
|
8Q2P
 
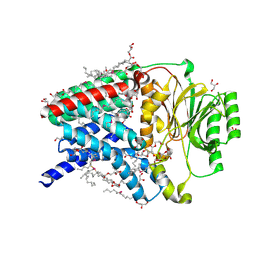 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli by using Se-MAG for the the lipid cubic phase crystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Apolipoprotein N-acyltransferase, ... | | Authors: | Huang, C.-Y, Boland, C, Kaki, S.S, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Se-MAG Is a Convenient Additive for Experimental Phasing and Structure Determination of Membrane Proteins Crystallised by the Lipid Cubic Phase (In Meso) Method
Crystals, 2023
|
|
8Q2O
 
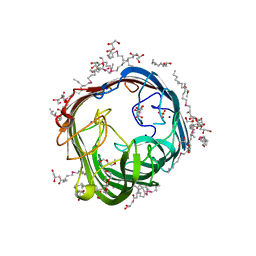 | | Structure of alginate transporter AlgE from P. aeruginosa PAO1 by using Se-MAG for the the lipid cubic phase crystallization | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alginate production protein AlgE, ... | | Authors: | Huang, C.-Y, Boland, C, Kaki, S.S, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Se-MAG Is a Convenient Additive for Experimental Phasing and Structure Determination of Membrane Proteins Crystallised by the Lipid Cubic Phase (In Meso) Method
Crystals, 2023
|
|
8B0K
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli (Apo form) | | Descriptor: | Apolipoprotein N-acyltransferase | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0O
 
 | | Cryo-EM structure apolipoprotein N-acyltransferase Lnt from E.coli in complex with FP3 | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{R})-3-[(2~{R})-3-[[(2~{R})-1-[[(2~{R})-1-[[(2~{R})-6-[(2-aminophenyl)carbonylamino]-1-azanyl-1-oxidanylidene-hexan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0L
 
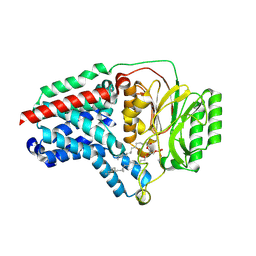 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0N
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{S})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-oxidanyl-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0P
 
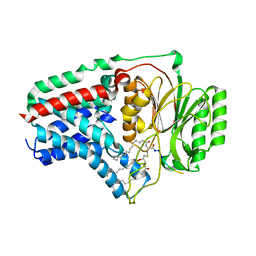 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Pam3 | | Descriptor: | Apolipoprotein N-acyltransferase, Pam3-SKKKK, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8B0M
 
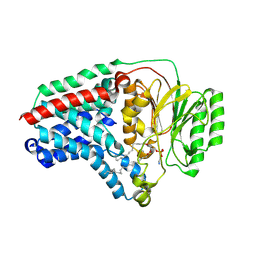 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with PE (C387S mutant) | | Descriptor: | Apolipoprotein N-acyltransferase, PHOSPHATIDYLETHANOLAMINE | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
4O9R
 
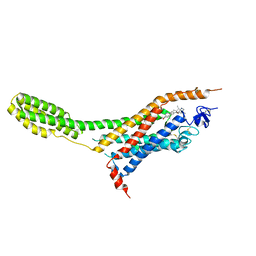 | | Human Smoothened Receptor structure in complex with cyclopamine | | Descriptor: | Cyclopamine, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Weierstall, U, James, D, White, T.A, Wang, D, Liu, W, Spence, J.C.H, Doak, R.B, Nelson, G, Fromme, P, Fromme, R, Grotjohann, I, Kupitz, C, Zatsepin, N.A, Liu, H, Basu, S, Wacker, D, Han, G.W, Katritch, V, Boutet, S, Messerschmidt, M, Willams, G.J, Koglin, J.E, Seibert, M.M, Klinker, M, Gati, C, Shoeman, R.L, Barty, A, Chapman, H.N, Kirian, R.A, Beyerlein, K.R, Stevens, R.C, Li, D, Shah, S.T.A, Howe, N, Caffrey, M, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography.
Nat Commun, 5, 2014
|
|
5DWK
 
 | | Diacylglycerol Kinase solved by multi crystal multi orientation native SAD | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | Authors: | Weinert, T, Olieric, V, Finke, A.D, Li, D, Caffrey, M, Wang, M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Data-collection strategy for challenging native SAD phasing.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8AQ2
 
 | | In meso structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa covalently linked with TITC | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
8AQ3
 
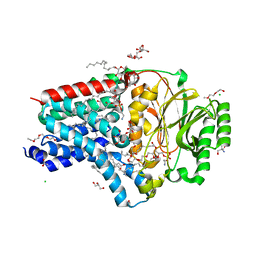 | | In surfo structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli in complex with PE | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
