6OQ5
 
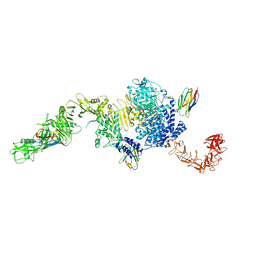 | | Structure of the full-length Clostridium difficile toxin B in complex with 3 VHHs | | Descriptor: | 5D, 7F, E3, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OQ7
 
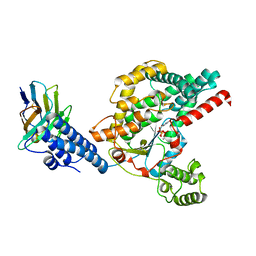 | | Structure of the GTD domain of Clostridium difficile toxin B in complex with VHH E3 | | Descriptor: | E3, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OQ6
 
 | |
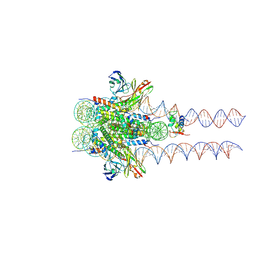
7K63
 
 | |
7K61
 
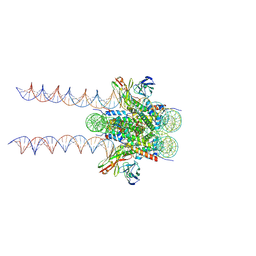 | | Cryo-EM structure of 197bp nucleosome aided by scFv | | Descriptor: | DNA (197-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.-R, Bai, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2020-11-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms.
Mol.Cell, 81, 2021
|
|
7K5X
 
 | |
7K5Y
 
 | |
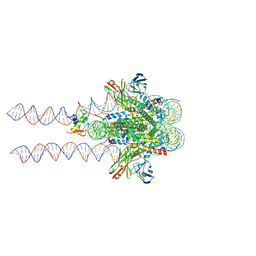
7K60
 
 | |
5O2E
 
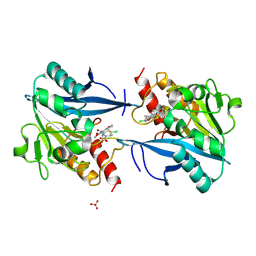 | | Crystal structure of NDM-1 in complex with hydrolyzed cefuroxime - new refinement | | Descriptor: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
6JBT
 
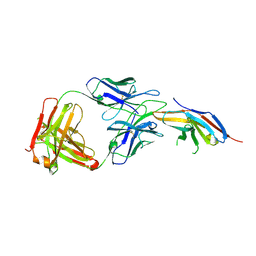 | | Complex structure of toripalimab-Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Guo, L, Tan, S, Chai, Y, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy.
Mabs, 11, 2019
|
|
8VFX
 
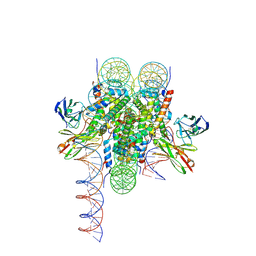 | | Cryo-EM structure of 186bp ALBN1 nucleosome aided by scFv | | Descriptor: | DNA (158-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.R, Bai, Y. | | Deposit date: | 2023-12-22 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.
Mol.Cell, 84, 2024
|
|
8VFY
 
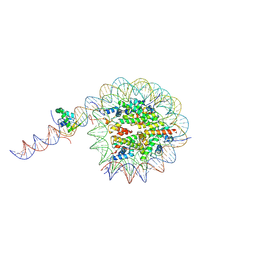 | |
8VG0
 
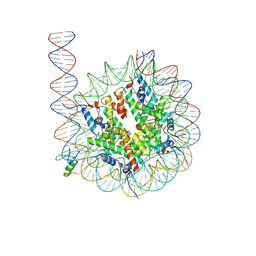 | | Cryo-EM structure of GATA4 in complex with ALBN1 nucleosome | | Descriptor: | DNA (159-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.R, Bai, Y. | | Deposit date: | 2023-12-22 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.
Mol.Cell, 84, 2024
|
|
5O2F
 
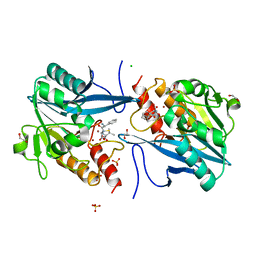 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
8VG1
 
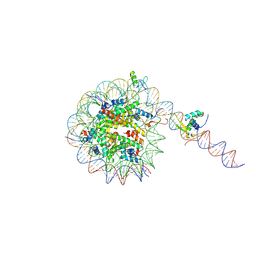 | |
8VFZ
 
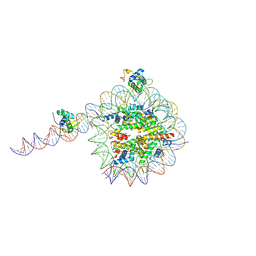 | |
9CB5
 
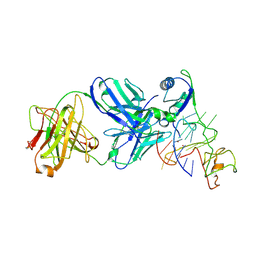 | | Crystal structure of nucleolin in complex with MYC promoter G-quadruplex | | Descriptor: | Fab heavy chain, Fab light chain, MYC promoter G-quadruplex, ... | | Authors: | Chen, L, Dickerhoff, J, Noinaj, N, Yang, D. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for nucleolin recognition of MYC promoter G-quadruplex.
Science, 388, 2025
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
5WCU
 
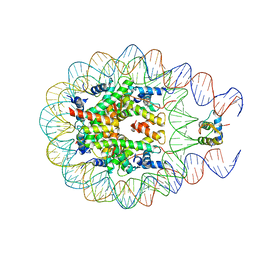 | |
4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | Descriptor: | CITRIC ACID, DNA (167-mer), H5, ... | | Authors: | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
8A8N
 
 | |
3VPI
 
 | |
3VPJ
 
 | |
7C88
 
 | | Complex structure of JS003 and PD-L1 | | Descriptor: | JS003 Heavy chain, JS003 Light chain, Programmed cell death 1 ligand 1 | | Authors: | Bi, X, Shi, R, Chai, Y, Qi, J, Yan, J, Tan, S. | | Deposit date: | 2020-05-29 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Identification of a hotspot on PD-L1 for pH-dependent binding by monoclonal antibodies for tumor therapy.
Signal Transduct Target Ther, 5, 2020
|
|
6J9M
 
 | | NmeBH+AcrIIC2 | | Descriptor: | AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
