5HVA
 
 | | Crystal structure of DR2231 in complex with dUMPNPP and magnesium. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DR2231, MAGNESIUM ION | | Authors: | Mota, C.S, Goncalves, A.M.D, de Sanctis, D. | | Deposit date: | 2016-01-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP.
FEBS J., 283, 2016
|
|
5HYL
 
 | | Crystal structure of DR2231_E47A mutant in complex with dUMPNPP and magnesium | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DR2231, MAGNESIUM ION | | Authors: | Mota, C.S, Goncalves, A.M.D, de Sanctis, D. | | Deposit date: | 2016-02-01 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP.
FEBS J., 283, 2016
|
|
4BLB
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLD
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI3p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, TRANSCRIPTIONAL ACTIVATOR GLI3, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5HWU
 
 | | Crystal Structure of DR2231_E46A mutant in complex with dUMPNPP and Manganese | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DR2231, GLYCEROL, ... | | Authors: | Mota, C.S, Goncalves, A.M.D, de Sanctis, D. | | Deposit date: | 2016-01-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deinococcus radiodurans DR2231 is a two-metal-ion mechanism hydrolase with exclusive activity on dUTP.
FEBS J., 283, 2016
|
|
5I0J
 
 | |
5I0M
 
 | |
5HZZ
 
 | |
5II6
 
 | | Crystal structure of the ZP-N1 domain of mouse sperm receptor ZP2 at 0.95 A resolution | | Descriptor: | Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Han, L, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5IIB
 
 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.64 A resolution (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Egg-lysin, ... | | Authors: | Raj, I, Sadat Al-Hosseini, H, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5II8
 
 | | Orthorhombic crystal structure of red abalone lysin at 0.99 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5IIA
 
 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.7 A resolution (crystal form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Egg-lysin, Vitelline envelope sperm lysin receptor | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
4BAG
 
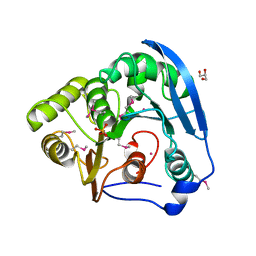 | |
4C0W
 
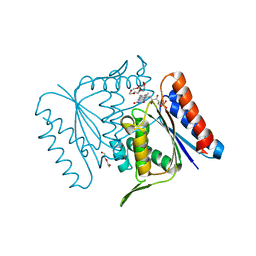 | |
4C14
 
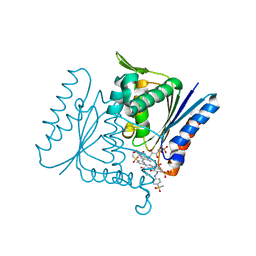 | | The crystal strucuture of PpAzoR in complex with reactive black 5 (RB5) | | Descriptor: | DODECAETHYLENE GLYCOL, FMN-DEPENDENT NADH-AZOREDUCTASE 1, [5-[3-[2-[[4-[2-[1-azanyl-7-[2-[4-[methyl-bis(oxidanyl)-$l^{4}-sulfanyl]phenyl]hydrazinyl]-8-oxidanyl-3,6-bis[tris(oxidanyl)-$l^{4}-sulfanyl]naphthalen-2-yl]hydrazinyl]phenyl]-bis(oxidanyl)-$l^{4}-sulfanyl]ethoxy]-7,8-dimethyl-2,4-bis(oxidanylidene)benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Goncalves, A.M.D, de Sanctis, D, Bento, I. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of Pseudomonas putida azoreductase - the active site revisited.
FEBS J., 280, 2013
|
|
4AD8
 
 | | Crystal structure of a deletion mutant of Deinococcus radiodurans RecN | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.998 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4C0X
 
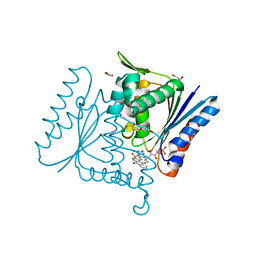 | | The crystal strucuture of PpAzoR in complex with anthraquinone-2- sulfonate | | Descriptor: | 9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN-DEPENDENT NADH-AZOREDUCTASE 1, ... | | Authors: | Goncalves, A.M.D, de Sanctis, D, Bento, I. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The Crystal Structure of Pseudomonas Putida Azor: The Active Site Revisited.
FEBS J., 280, 2013
|
|
4BAJ
 
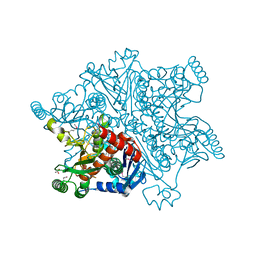 | | MYCOBACTERIUM TUBERCULOSIS CHORISMATE SYNTHASE after exposure to 266nm UV laser | | Descriptor: | ACETATE ION, CHORISMATE SYNTHASE | | Authors: | Pereira, P.J.B, Royant, A, Panjikar, S, de Sanctis, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In-house UV radiation-damage-induced phasing of selenomethionine-labeled protein structures.
J. Struct. Biol., 181, 2013
|
|
4AWE
 
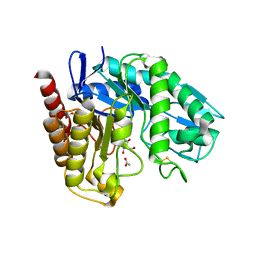 | | The Crystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goncalves, A.M.D, Silva, C.S, De Sanctis, D, Bento, I. | | Deposit date: | 2012-06-01 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Endo-Beta-D-1,4-Mannanase from Chrysonilia Sitophila Displays a Novel Loop Arrangement for Substrate Selectivity
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4ABX
 
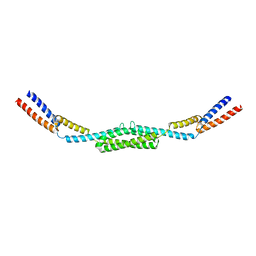 | | Crystal structure of Deinococcus radiodurans RecN coiled-coil domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4ABY
 
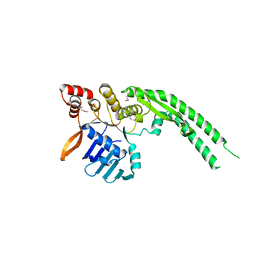 | | Crystal structure of Deinococcus radiodurans RecN head domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4BAI
 
 | | Mycobacterium tuberculosis Chorismate synthase before exposure to 266 nm UV laser | | Descriptor: | ACETATE ION, CHORISMATE SYNTHASE | | Authors: | Pereira, P.J.B, Royant, A, Panjikar, S, de Sanctis, D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In-house UV radiation-damage-induced phasing of selenomethionine-labeled protein structures.
J. Struct. Biol., 181, 2013
|
|
2X8S
 
 | | Crystal Structure of the Abn2 D171A mutant in complex with arabinotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | deSanctis, D, Inacio, J.M, Lindley, P.F, de Sa-Nogueira, I, Bento, I. | | Deposit date: | 2010-03-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Evidence for the Role of Calcium in the Glycosidase Reaction of Gh43 Arabinanases.
FEBS J., 277, 2010
|
|
2X8T
 
 | | Crystal Structure of the Abn2 H318A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | deSanctis, D, Inacio, J.M, Lindley, P.F, de Sa-Nogueira, I, Bento, I. | | Deposit date: | 2010-03-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | New Evidence for the Role of Calcium in the Glycosidase Reaction of Gh43 Arabinanases.
FEBS J., 277, 2010
|
|
7PFP
 
 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
