5OH2
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Pyrrolidin-2-one (Butyrolactam) | | Descriptor: | Cereblon isoform 4, ZINC ION, pyrrolidin-2-one | | Authors: | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2017-07-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
5T87
 
 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
7MHU
 
 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
6FIB
 
 | | Structure of human 4-1BB ligand | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor ligand superfamily member 9,4-1BBL -CH/CL fusion, Tumor necrosis factor ligand superfamily member 9,Uncharacterized protein | | Authors: | Joseph, C, Claus, C, Ferrara, C, von Hirschheydt, T, Prince, C, Funk, D, Klein, C, Benz, J. | | Deposit date: | 2018-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tumor-targeted 4-1BB agonists for combination with T cell bispecific antibodies as off-the-shelf therapy.
Sci Transl Med, 11, 2019
|
|
6HQV
 
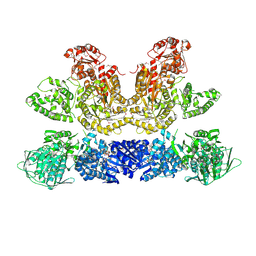 | | Pentafunctional AROM Complex from Chaetomium thermophilum | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, GLUTAMIC ACID, ... | | Authors: | Arora Verasto, H, Hartmann, M.D. | | Deposit date: | 2018-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architecture and functional dynamics of the pentafunctional AROM complex.
Nat.Chem.Biol., 16, 2020
|
|
3DA4
 
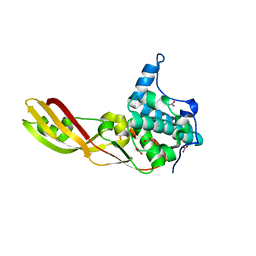 | | Crystal Structure of Colicin M, a Novel Phosphatase Specifically Imported by Escherichia Coli | | Descriptor: | Colicin-M, NITRATE ION | | Authors: | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | Deposit date: | 2008-05-28 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
1P9X
 
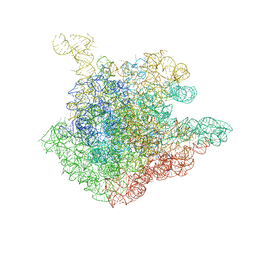 | | THE CRYSTAL STRUCTURE OF THE 50S LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS COMPLEXED WITH TELITHROMYCIN KETOLIDE ANTIBIOTIC | | Descriptor: | 23S RIBOSOMAL RNA, TELITHROMYCIN | | Authors: | Berisio, R, Harms, J, Schluenzen, F, Zarivach, R, Hansen, H.A, Fucini, P, Yonath, A. | | Deposit date: | 2003-05-13 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into the antibiotic action of telithromycin against resistant mutants
J.Bacteriol., 185, 2003
|
|
1OND
 
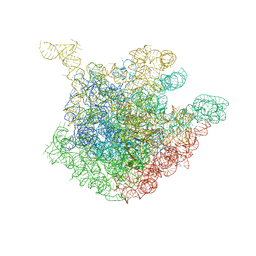 | | THE CRYSTAL STRUCTURE OF THE 50S LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS COMPLEXED WITH TROLEANDOMYCIN MACROLIDE ANTIBIOTIC | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L22, 50S ribosomal protein L32, ... | | Authors: | Berisio, R, Schluenzen, F, Harms, J, Bashan, A, Auerbach, T, Baram, D, Yonath, A. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into the role of the ribosomal tunnel in cellular regulation
Nat.Struct.Biol., 10, 2003
|
|
6T76
 
 | | PII-like protein CutA from Nostoc sp. PCC 7120 in apo form | | Descriptor: | Periplasmic divalent cation tolerance protein, SULFATE ION | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
6T7E
 
 | | PII-like protein CutA from Nostoc sp. PCC7120 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Periplasmic divalent cation tolerance protein | | Authors: | Selim, K.A, Albrecht, R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 288, 2021
|
|
1NKW
 
 | | Crystal Structure Of The Large Ribosomal Subunit From Deinococcus Radiodurans | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Harms, J.M, Schluenzen, F, Zarivach, R, Bashan, A, Gat, S, Agmon, I, Bartels, H, Franceschi, F, Yonath, A. | | Deposit date: | 2003-01-05 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High resolution structure of the large ribosomal subunit from a mesophilic eubacterium
Cell(Cambridge,Mass.), 107, 2001
|
|
8CMP
 
 | |
3DA3
 
 | | Crystal Structure of Colicin M, A Novel Phosphatase Specifically Imported by Escherichia Coli | | Descriptor: | Colicin-M, MAGNESIUM ION | | Authors: | Zeth, K, Albrecht, R, Romer, C, Braun, V. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of colicin M, a novel phosphatase specifically imported by Escherichia coli
J.Biol.Chem., 283, 2008
|
|
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2H
 
 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
2YJJ
 
 | |
2YJK
 
 | |
8BC7
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex an aminoglutarimide degron peptide | | Descriptor: | Cereblon isoform 4, PHE-PHE-GLU-GLN-MET-GLN-QCI, S-Thalidomide, ... | | Authors: | Heim, C, Albrecht, R, Spring, A.K, Hartmann, M.D. | | Deposit date: | 2022-10-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Identification and structural basis of C-terminal cyclic imides as natural degrons for cereblon.
Biochem.Biophys.Res.Commun., 637, 2022
|
|
6H9M
 
 | | Coiled-coil domain-containing protein 90B residues 43-125 from Homo sapiens fused to a GCN4 adaptor | | Descriptor: | Coiled-coil domain-containing protein 90B, mitochondrial,General control protein GCN4 | | Authors: | Adlakha, J, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | Deposit date: | 2018-08-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of MCU-Binding Proteins MCUR1 and CCDC90B - Representatives of a Protein Family Conserved in Prokaryotes and Eukaryotic Organelles.
Structure, 27, 2019
|
|
3TQ2
 
 | |
9EZZ
 
 | | Bacterial histone protein HBb from Bdellovibrio bacteriovorus bound to DNA | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*CP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hu, Y, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2024-04-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacterial histone HBb from Bdellovibrio bacteriovorus compacts DNA by bending.
Nucleic Acids Res., 52, 2024
|
|
9F0E
 
 | |
9F2C
 
 | |
2WQ1
 
 | |
