1NWI
 
 | |
1YHU
 
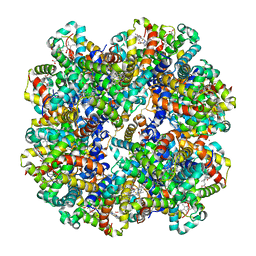 | | Crystal structure of Riftia pachyptila C1 hemoglobin reveals novel assembly of 24 subunits. | | Descriptor: | Giant hemoglobins B chain, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Flores, J.F, Fisher, C.R, Carney, S.L, Green, B.N, Freytag, J.K, Schaeffer, S.W, Royer, W.E. | | Deposit date: | 2005-01-10 | | Release date: | 2005-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Sulfide binding is mediated by zinc ions discovered in the crystal structure of a hydrothermal vent tubeworm hemoglobin.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
9BWR
 
 | | X-ray Counterpart to the Neutron Structure of Reduced Tyr34Phe MnSOD | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Cone, E.A, Dasgupta, M, Lutz, W.E, Kumar, S, Natarajan, A, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-05-21 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The role of Tyr34 in proton coupled electron transfer and product inhibition of manganese superoxide dismutase.
Nat Commun, 16, 2025
|
|
9BWQ
 
 | | X-ray Counterpart to the Neutron Structure of Peroxide-Soaked Tyr34Phe MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Cone, E.A, Dasgupta, M, Lutz, W.E, Kumar, S, Natarajan, A, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-05-21 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The role of Tyr34 in proton coupled electron transfer and product inhibition of manganese superoxide dismutase.
Nat Commun, 16, 2025
|
|
9BWM
 
 | | Neutron Structure of Oxidized Tyr34Phe MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Cone, E.A, Dasgupta, M, Lutz, W.E, Kumar, S, Natarajan, A, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-05-21 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (2.28 Å) | | Cite: | The role of Tyr34 in proton coupled electron transfer and product inhibition of manganese superoxide dismutase.
Nat Commun, 16, 2025
|
|
4LCJ
 
 | | CtBP2 in complex with substrate MTOB | | Descriptor: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | Deposit date: | 2013-06-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
9B2G
 
 | | Crystal structure of short chain dehydrogenase reductase 9 (short chain dehydrogenase reductase 9C4) in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dehydrogenase/reductase SDR family member 9 | | Authors: | Pakhomova, S, Belyaeva, O.V, Boeglin, W.E, Kedishvili, N.Y, Brash, A.R, Newcomer, M.E, Popov, K.M. | | Deposit date: | 2024-03-15 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The large substrate binding pocket of dehydrogenase reductase 9 underlies its ability to oxidize diverse pro-inflammatory and pro-resolving oxylipins.
To Be Published
|
|
9B2F
 
 | | Crystal structure of short chain dehydrogenase reductase 9 (short chain dehydrogenase reductase 9C4) in complex with NAD+ | | Descriptor: | Dehydrogenase/reductase SDR family member 9, MALONATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pakhomova, S, Belyaeva, O.V, Boeglin, W.E, Kedishvili, N.Y, Brash, A.R, Newcomer, M.E, Popov, K.M. | | Deposit date: | 2024-03-15 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.662 Å) | | Cite: | The large substrate binding pocket of dehydrogenase reductase 9 underlies its ability to oxidize diverse pro-inflammatory and pro-resolving oxylipins.
To Be Published
|
|
9BVY
 
 | | Neutron Structure of Peroxide-Soaked Tyr34Phe MnSOD | | Descriptor: | HYDROGEN PEROXIDE, MANGANESE (II) ION, Superoxide dismutase [Mn], ... | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Cone, E.A, Dasgupta, M, Lutz, W.E, Kumar, S, Natarajan, A, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-05-20 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | The role of Tyr34 in proton coupled electron transfer and product inhibition of manganese superoxide dismutase.
Nat Commun, 16, 2025
|
|
9BW2
 
 | | Neutron Structure of Reduced Tyr34Phe MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Azadmanesh, J, Slobodnik, K, Struble, L.R, Cone, E.A, Dasgupta, M, Lutz, W.E, Kumar, S, Natarajan, A, Coates, L, Weiss, K.L, Myles, D.A.A, Kroll, T, Borgstahl, G.E.O. | | Deposit date: | 2024-05-20 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (2.5 Å) | | Cite: | The role of Tyr34 in proton coupled electron transfer and product inhibition of manganese superoxide dismutase.
Nat Commun, 16, 2025
|
|
1J6X
 
 | | CRYSTAL STRUCTURE OF HELICOBACTER PYLORI LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1GND
 
 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Authors: | Schalk, I, Zeng, K, Wu, S.-K, Stura, E.A, Metteson, J, Huang, M, Tandon, A, Wilson, I.A, Balch, W.E. | | Deposit date: | 1996-07-10 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and mutational analysis of Rab GDP-dissociation inhibitor.
Nature, 381, 1996
|
|
1INN
 
 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, P21 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1J6W
 
 | | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1J6V
 
 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, C2 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
4YDF
 
 | | Crystal structure of compound 9 in complex with HTLV-1 Protease | | Descriptor: | HTLV-1 Protease, N-benzyl-N-[(3S,4S)-4-{benzyl[(4-nitrophenyl)sulfonyl]amino}pyrrolidin-3-yl]-3-nitrobenzenesulfonamide, SULFATE ION | | Authors: | Kuhnert, M, Blum, A, Steuber, H, Diederich, W.E. | | Deposit date: | 2015-02-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Privileged Structures Meet Human T-Cell Leukemia Virus-1 (HTLV-1): C2-Symmetric 3,4-Disubstituted Pyrrolidines as Nonpeptidic HTLV-1 Protease Inhibitors.
J.Med.Chem., 58, 2015
|
|
4ZF8
 
 | | Cytochrome P450 pentamutant from BM3 with bound Metyrapone | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, METYRAPONE, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.766 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
5KEG
 
 | | Crystal structure of APOBEC3A in complex with a single-stranded DNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*TP*TP*CP*TP*T)-3'), ... | | Authors: | Kouno, T, Hilbert, B.J, Silvas, T, Royer, W.E, Matsuo, H, Schiffer, C.A. | | Deposit date: | 2016-06-09 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity.
Nat Commun, 8, 2017
|
|
4YDG
 
 | | Crystal structure of compound 10 in complex with HTLV-1 Protease | | Descriptor: | HTLV-1 protease, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(4-AMINO-N-BENZYLBENZENESULFONAMIDE), SULFATE ION | | Authors: | Kuhnert, M, Blum, A, Steuber, H, Diederich, W.E. | | Deposit date: | 2015-02-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Privileged Structures Meet Human T-Cell Leukemia Virus-1 (HTLV-1): C2-Symmetric 3,4-Disubstituted Pyrrolidines as Nonpeptidic HTLV-1 Protease Inhibitors.
J.Med.Chem., 58, 2015
|
|
5JRM
 
 | | Crystal Structure of a Xylanase at 1.56 Angstroem resolution | | Descriptor: | Endo-1,4-beta-xylanase, GLYCEROL, SULFATE ION | | Authors: | Gomez, S, Payne, A.M, Savko, M, Fox, G.C, Shepard, W.E, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and functional characterization of a highly stable endo-beta-1,4-xylanase from Fusarium oxysporum and its development as an efficient immobilized biocatalyst.
Biotechnol Biofuels, 9, 2016
|
|
4ZFB
 
 | | Cytochrome P450 pentamutant from BM3 bound to Palmitic Acid | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
4ZFA
 
 | | Cytochrome P450 wild type from BM3 with bound PEG | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.765 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
4ZF6
 
 | | Cytochrome P450 pentamutant from BM3 with bound PEG | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional P-450/NADPH-P450 reductase, NICKEL (II) ION, ... | | Authors: | Rogers, W.E, Othman, T, Heidary, D.K, Huxford, T. | | Deposit date: | 2015-04-21 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Effect of Mutation and Substrate Binding on the Stability of Cytochrome P450BM3 Variants.
Biochemistry, 55, 2016
|
|
5LAC
 
 | |
5LAK
 
 | | Ligand-bound structure of Cavally Virus 3CL Protease | | Descriptor: | 3Cl Protease, BEZ-TYR-TYR-ASN-ECC Peptide inhibitor, PENTAETHYLENE GLYCOL, ... | | Authors: | Kanitz, M, Heine, A, Diederich, W.E. | | Deposit date: | 2016-06-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus.
Virology, 533, 2019
|
|
