8WPF
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exogenous DNA bearing one abasic site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, A22R DNA polymerase processivity factor, DNA polymerase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPK
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exgenous DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPE
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 (tag-free A22) with exogenous DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, A22R DNA polymerase processivity factor, DNA polymerase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPP
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with endogenous DNA | | Descriptor: | A22R DNA polymerase processivity factor, DNA polymerase, E4R Uracil-DNA glycosylase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
5WUA
 
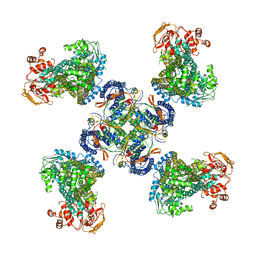 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
5XF8
 
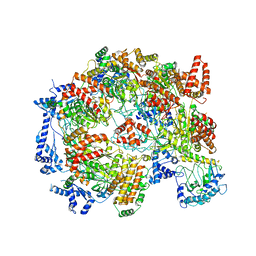 | | Cryo-EM structure of the Cdt1-MCM2-7 complex in AMPPNP state | | Descriptor: | Cell division cycle protein CDT1, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Zhai, Y, Cheng, E, Wu, H, Li, N, Yung, P.Y, Gao, N, Tye, B.K. | | Deposit date: | 2017-04-09 | | Release date: | 2017-05-03 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer
Nat. Struct. Mol. Biol., 24, 2017
|
|
5H4P
 
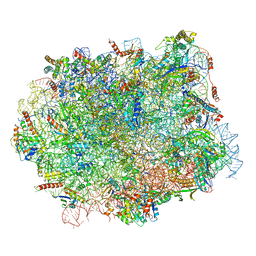 | | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Ma, C, Wu, S, Li, N, Chen, Y, Yan, K, Li, Z, Zheng, L, Lei, J, Woolford, J.L, Gao, N. | | Deposit date: | 2016-11-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1
Nat. Struct. Mol. Biol., 24, 2017
|
|
5H5U
 
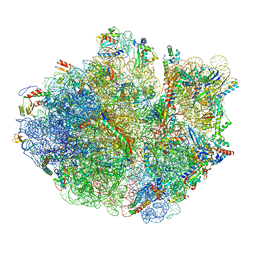 | | Mechanistic insights into the alternative translation termination by ArfA and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ma, C, Kurita, D, Li, N, Chen, Y, Himeno, H, Gao, N. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanistic insights into the alternative translation termination by ArfA and RF2
Nature, 541, 2017
|
|
8IB2
 
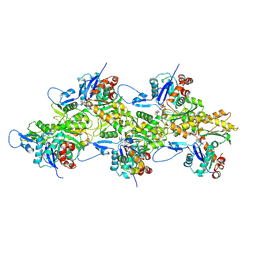 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, Pointed-end segment, headpiece domain of dematin optimized | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
8IAH
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State I, Global map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
8IAI
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State II, Global map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
3PN1
 
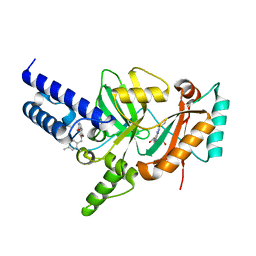 | | Novel Bacterial NAD+-dependent DNA Ligase Inhibitors with Broad Spectrum Potency and Antibacterial Efficacy In Vivo | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-(butylsulfanyl)adenosine, DNA ligase | | Authors: | Mills, S, Eakin, A, Buurman, E, Newman, J, Gao, N, Huynh, H, Johnson, K, Lahiri, S, Shapiro, A, Walkup, G, Wei, Y, Stokes, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Bacterial NAD+-Dependent DNA Ligase Inhibitors with Broad-Spectrum Activity and Antibacterial Efficacy In Vivo.
Antimicrob.Agents Chemother., 55, 2011
|
|
4FN7
 
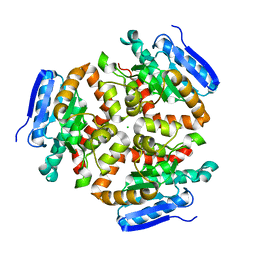 | | Apo Structure of the Mtb enoyol CoA isomerase (Rv0632c) | | Descriptor: | CHLORIDE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-19 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | "Crystal Structure and Mechanism of the Prokaryotic Enoyl-CoA Isomerase (ECI)"
To be Published
|
|
4FND
 
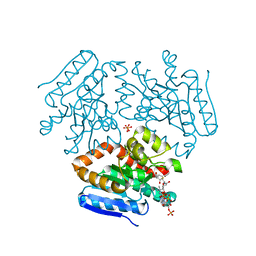 | | Crystal structure of the Mtb enoyl CoA isomerase in complex with hydroxyhexanoyl CoA | | Descriptor: | (S)-3-Hydroxyhexanoyl-CoA, Enoyl-CoA hydratase/isomerase family protein, SULFATE ION | | Authors: | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-19 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and mechanism of the prokaryotic enoyl CoA isomerase
To be Published
|
|
4FN8
 
 | | Crystal structure of the Mtb enoyl CoA isomerase (Rv0632c)in complex with acetoacetyl CoA | | Descriptor: | ACETOACETYL-COENZYME A, Enoyl-CoA hydratase/isomerase family protein, SULFATE ION | | Authors: | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-19 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | Crystal structure and mechanism of the prokaryotic enoyl CoA isomerase (ECI)
To be Published
|
|
4FNB
 
 | | Crystal structure of the Mtb enoyl CoA isomerase (Rv0632c) in complex with hydroxybutyrl CoA | | Descriptor: | 3-HYDROXYBUTANOYL-COENZYME A, Enoyl-CoA hydratase/isomerase family protein, SULFATE ION | | Authors: | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-19 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Mechanism of the Prokaryotic Enoyl CoA Isomerase
To be Published
|
|
7V3V
 
 | | Cryo-EM structure of MCM double hexamer bound with DDK in State I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 7, DDK kinase regulatory subunit DBF4, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7W8G
 
 | | Cryo-EM structure of MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Tye, B, Zhai, Y, Gao, N. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7V3U
 
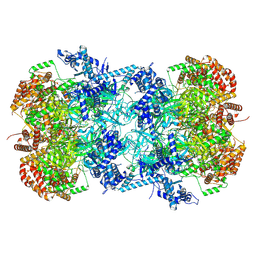 | | Cryo-EM structure of MCM double hexamer with structured Mcm4-NSD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7XBK
 
 | | Structure and mechanism of a mitochondrial AAA+ disaggregase CLPB | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform 2 of Caseinolytic peptidase B protein homolog, MAGNESIUM ION, ... | | Authors: | Wu, D, Liu, Y, Dai, Y, Wang, G, Lu, G, Chen, Y, Li, N, Lin, J, Gao, N. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Comprehensive structural characterization of the human AAA+ disaggregase CLPB in the apo- and substrate-bound states reveals a unique mode of action driven by oligomerization.
Plos Biol., 21, 2023
|
|
7XFN
 
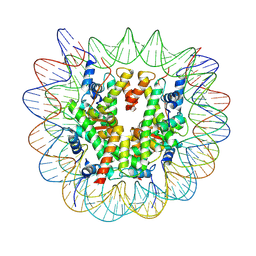 | | Structure of nucleosome-DI complex (-55I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFI
 
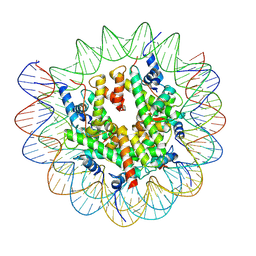 | | Structure of nucleosome-DI complex (-50I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFH
 
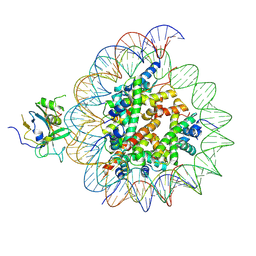 | | Structure of nucleosome-AAG complex (A-30I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFM
 
 | | Structure of nucleosome-AAG complex (A-53I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
7XFC
 
 | | Structure of nucleosome-DI complex (-30I, Apo state) | | Descriptor: | DNA (152-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
