7SNF
 
 | | Structure of G6PD-WT dimer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3OUQ
 
 | | Structure of N-terminal hexaheme fragment of GSU1996 | | Descriptor: | Cytochrome c family protein, HEME C | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12nm protein with interacting hemes.
J.Struct.Biol., 174, 2011
|
|
3OV0
 
 | | Structure of dodecaheme cytochrome c GSU1996 | | Descriptor: | Cytochrome c family protein, HEME C | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12nm protein with interacting hemes.
J.Struct.Biol., 174, 2011
|
|
7BFZ
 
 | | X-ray structure of human prostate-specific membrane antigen(PSMA) in complex with a inhibitor Glu-490 | | Descriptor: | (((S)-1-carboxy-5-((E)-2-cyano-3-(5-(1-(3-methoxy-3-oxopropyl)-1,2,3,4-tetrahydroquinolin-6-yl)thiophen-2-yl)acrylamido)pentyl)carbamoyl)-L-glutamic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rakhimbekova, A, Motlova, L, Barinka, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A prostate-specific membrane antigen activated molecular rotor for real-time fluorescence imaging.
Nat Commun, 12, 2021
|
|
3OUE
 
 | | Structure of C-terminal hexaheme fragment of GSU1996 | | Descriptor: | Cytochrome c family protein, HEME C, SULFATE ION | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12nm protein with interacting hemes.
J.Struct.Biol., 174, 2011
|
|
7WIX
 
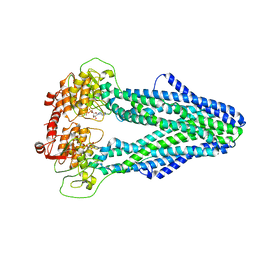 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIV
 
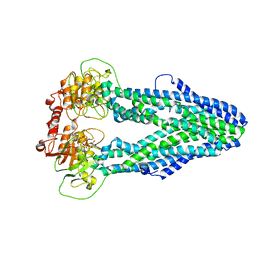 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in complex with an AMP-PNP | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIU
 
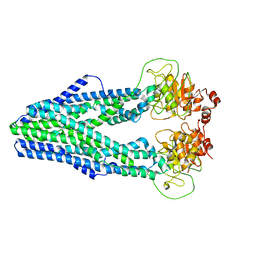 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB in inward-facing state | | Descriptor: | Mycobactin import ATP-binding/permease protein IrtA, Mycobactin import ATP-binding/permease protein IrtB | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7WIW
 
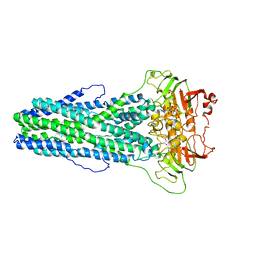 | | Cryo-EM structure of Mycobacterium tuberculosis irtAB complexed with ATP in an occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mycobactin import ATP-binding/permease protein IrtA, ... | | Authors: | Zhang, B, Sun, S, Yang, H, Rao, Z. | | Deposit date: | 2022-01-05 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures for the Mycobacterium tuberculosis iron-loaded siderophore transporter IrtAB.
Protein Cell, 14, 2023
|
|
7UAL
 
 | |
7UC2
 
 | |
7FDK
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to mACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDH
 
 | | SARS-COV-2 Spike RBDMACSp25 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDI
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDG
 
 | | SARS-COV-2 Spike RBDMACSp6 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
5HGZ
 
 | | Crystal structure of human Naa60 in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, MALONIC ACID, N-alpha-acetyltransferase 60 | | Authors: | Chen, J.Y, Liu, L, Yun, C.H. | | Deposit date: | 2016-01-09 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Structure and function of human Naa60 (NatF), a Golgi-localized bi-functional acetyltransferase
Sci Rep, 6, 2016
|
|
5HH1
 
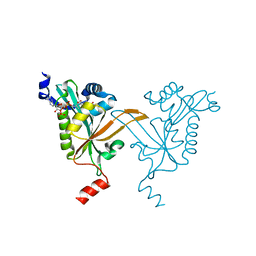 | |
5HH0
 
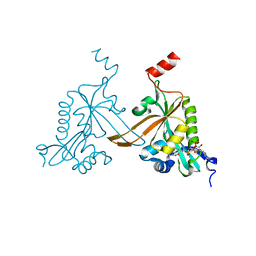 | |
3UO8
 
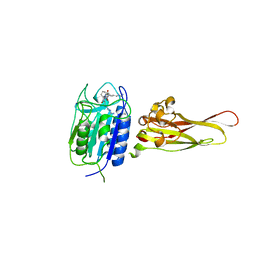 | | Crystal structure of the MALT1 paracaspase (P1 form) | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1, Z-Val-Arg-Pro-DL-Arg-fluoromethylketone | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4RBY
 
 | |
4RC0
 
 | |
4R0E
 
 | |
3V86
 
 | |
3UOA
 
 | | Crystal structure of the MALT1 paracaspase (P21 form) | | Descriptor: | MAGNESIUM ION, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, Z-Val-Arg-Pro-DL-Arg-fluoromethylketone | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7E9O
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
