3OF5
 
 | | Crystal Structure of a Dethiobiotin Synthetase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, Dethiobiotin synthetase, SODIUM ION | | Authors: | Brunzelle, J.S, Skarina, T, Gordon, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-13 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of a Dethiobiotin Synthetase from Francisella tularensis subsp. tularensis SCHU S4
TO BE PUBLISHED
|
|
3OYT
 
 | | 1.84 Angstrom resolution crystal structure of 3-oxoacyl-(acyl carrier protein) synthase I (fabB) from Yersinia pestis CO92 | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase I, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-23 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | 1.84 Angstrom resolution crystal structure of 3-oxoacyl-(acyl carrier protein) synthase I (fabB) from Yersinia pestis CO92
To be Published
|
|
3P7M
 
 | | Structure of putative lactate dehydrogenase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Malate dehydrogenase, PHOSPHATE ION | | Authors: | Osinski, T, Cymborowski, M, Zimmerman, M.D, Gordon, E, Grimshaw, S, Skarina, T, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of putative lactate dehydrogenase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3OJC
 
 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
4I3F
 
 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
2M2J
 
 | | Solution NMR structure of the N-terminal domain of STM1478 from Salmonella typhimurium LT2: Target STR147A of the Northeast Structural Genomics consortium (NESG), and APC101565 of the Midwest Center for Structural Genomics (MCSG). | | Descriptor: | Putative periplasmic protein | | Authors: | Houliston, S, Yee, A, Lemak, A, Garcia, M, Wu, B, Savchenko, A, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-21 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
4EJ7
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside 3'-phosphotransferase AphA1-IAB, CALCIUM ION, ... | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Evdokimova, E, Egorova, O, Di Leo, R, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4EW5
 
 | | C-terminal domain of inner membrane protein CigR from Salmonella enterica. | | Descriptor: | 1,2-ETHANEDIOL, CigR Protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Niemann, G.S, Merkley, E.D, Savchenko, A, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | C-terminal domain of inner membrane protein CigR from Salmonella enterica.
To be Published
|
|
3H02
 
 | | 2.15 Angstrom Resolution Crystal Structure of Naphthoate Synthase from Salmonella typhimurium. | | Descriptor: | BICARBONATE ION, Naphthoate synthase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Resolution Crystal Structure of Naphthoate Synthase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3H0P
 
 | | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium. | | Descriptor: | S-malonyltransferase, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal Structure of an Acyl Carrier Protein S-malonyltransferase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3H5Q
 
 | | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus | | Descriptor: | Pyrimidine-nucleoside phosphorylase, SULFATE ION, THYMIDINE | | Authors: | Shumilin, I.A, Zimmerman, M, Cymborowski, M, Skarina, T, Onopriyenko, O, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-26 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a putative pyrimidine-nucleoside phosphorylase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3GRI
 
 | | The Crystal Structure of a Dihydroorotase from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Dihydroorotase from Staphylococcus aureus
To be Published
|
|
4EUY
 
 | | Crystal structure of thioredoxin-like protein BCE_0499 from Bacillus cereus ATCC 10987 | | Descriptor: | Uncharacterized protein | | Authors: | Shabalin, I.G, Kagan, O, Chruszcz, M, Grabowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of thioredoxin-like protein BCE_0499 from
Bacillus cereus
To be Published
|
|
3GSD
 
 | | 2.05 Angstrom structure of a divalent-cation tolerance protein (CutA) from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Structure of a Divalent-cation Tolerance Protein (CutA) from Yersinia pestis
TO BE PUBLISHED
|
|
3SKS
 
 | | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames | | Descriptor: | PHOSPHATE ION, Putative Oligoendopeptidase F, ZINC ION | | Authors: | Wajerowicz, W, Onopriyenko, O, Porebski, P, Domagalski, M, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | Descriptor: | Effector protein hopAB3 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3H2Y
 
 | | Crystal structure of YqeH GTPase from Bacillus anthracis with dGDP bound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, GTPase family protein | | Authors: | Brunzelle, J.S, Anderson, S.M, Xu, X, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of YqeH GTPase from Bacillus anthracis with dGDP Bound
To be Published
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | Descriptor: | Effector protein hopAB1 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
4FEV
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor pyrazolopyrimidine PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
3T7B
 
 | | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis | | Descriptor: | Acetylglutamate kinase, GLUTAMIC ACID, S,R MESO-TARTARIC ACID | | Authors: | Demas, M.W, Solberg, R.G, Cooper, D.R, Chruszcz, M, Porebski, P.J, Zheng, H, Onopriyenko, O, Skarina, T, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis
To be Published
|
|
3HFR
 
 | | Crystal structure of glutamate racemase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Glutamate racemase, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Klimecka, M.M, Cymborowski, M, Skarina, T, Onopriyenko, O, Stam, J, Otwinowski, Z, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes
TO BE PUBLISHED
|
|
3T7Y
 
 | | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis | | Descriptor: | CHLORIDE ION, FORMIC ACID, SODIUM ION, ... | | Authors: | Singer, A.U, Wawrzak, Z, Skarina, T, Saikali, P, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis
TO BE PUBLISHED
|
|
4GKH
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
2M4E
 
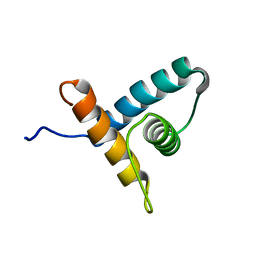 | | Solution NMR structure of VV2_0175 from Vibrio vulnificus, NESG target VnR1 and CSGID target IDP91333 | | Descriptor: | Putative uncharacterized protein | | Authors: | Wu, B, Yee, A, Houliston, S, Lemak, A, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of VV2_0175 from Vibrio vulnificus, NESG target VnR1 and CSGID target IDP91333
To be Published
|
|
3IAC
 
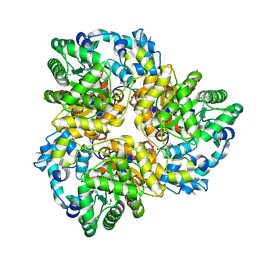 | | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium. | | Descriptor: | CHLORIDE ION, Glucuronate isomerase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium.
To be Published
|
|
