4FI9
 
 | | Structure of human SUN-KASH complex | | Descriptor: | Nesprin-2, SUN domain-containing protein 2 | | Authors: | Wang, W.J, Shi, Z.B. | | Deposit date: | 2012-06-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into SUN-KASH complexes across the nuclear envelope.
Cell Res., 22, 2012
|
|
4B45
 
 | |
3ZID
 
 | |
7F9W
 
 | | CD25 in complex with Fab | | Descriptor: | Heavy chain of Fab, Interleukin-2 receptor subunit alpha, Light chain of Fab | | Authors: | Liu, C. | | Deposit date: | 2021-07-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Two novel human anti-CD25 antibodies with antitumor activity inversely related to their affinity and in vitro activity.
Sci Rep, 11, 2021
|
|
6M79
 
 | |
7F0I
 
 | | phosphodiesterase-9A in complex with inhibitor 4b | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(6-fluoranyl-2-azaspiro[3.3]heptan-2-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, Isoform PDE9A2 of High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.70000887 Å) | | Cite: | Discovery of Potent Phosphodiesterase-9 Inhibitors for the Treatment of Hepatic Fibrosis
J.Med.Chem., 64, 2021
|
|
7QI8
 
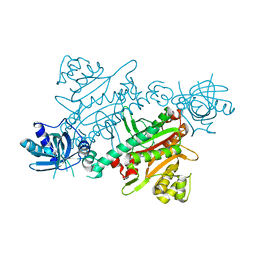 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE AND INHIBITOR | | Descriptor: | 2-azanyl-6-[(1~{S},7~{S})-2,2-bis(fluoranyl)-7-oxidanyl-cycloheptyl]-4-methoxy-7~{H}-pyrrolo[3,4-d]pyrimidin-5-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QHN
 
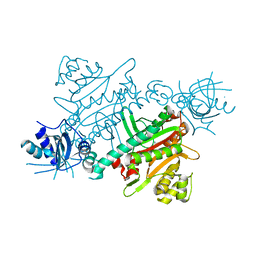 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE and an inhibitor | | Descriptor: | 6-azanyl-2-cyclohexyl-4-fluoranyl-1~{H}-pyrrolo[3,4-c]pyridin-3-one, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
7QH8
 
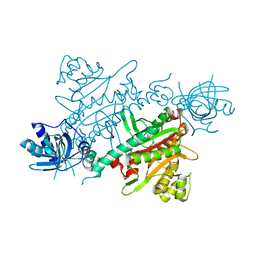 | | CRYSTAL STRUCTURE OF LYSYL-TRNA SYNTHETASE FROM Mycobacterium tuberculosis COMPLEXED WITH L-LYSINE | | Descriptor: | GLYCEROL, LYSINE, Lysine--tRNA ligase 1 | | Authors: | Dawson, A, Robinson, D.A, Tamjar, J, Wyatt, P, Green, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Lysyl-tRNA synthetase, a target for urgently needed M. tuberculosis drugs.
Nat Commun, 13, 2022
|
|
1LQF
 
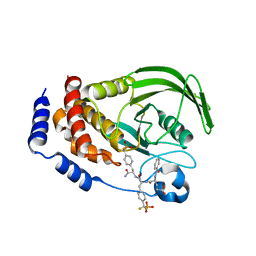 | | Structure of PTP1b in Complex with a Peptidic Bisphosphonate Inhibitor | | Descriptor: | N-BENZOYL-L-GLUTAMYL-[4-PHOSPHONO(DIFLUOROMETHYL)]-L-PHENYLALANINE-[4-PHOSPHONO(DIFLUORO-METHYL)]-L-PHENYLALANINEAMIDE, protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Asante-Appiah, E, Patel, S, Dufresne, C, Scapin, G. | | Deposit date: | 2002-05-10 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of PTP-1B in complex with a peptide inhibitor reveals an alternative binding mode for bisphosphonates.
Biochemistry, 41, 2002
|
|
3FF7
 
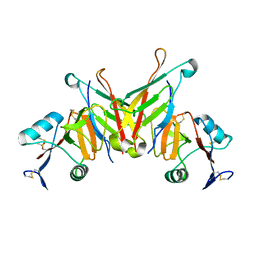 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | Descriptor: | ACETIC ACID, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
9JM0
 
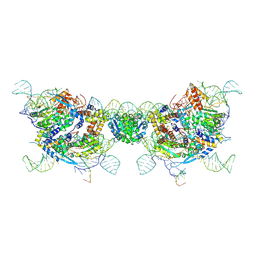 | | retron Ec86-effector fiber | | Descriptor: | DNA (85-MER), NICOTINAMIDE, RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*GP*UP*GP*CP*GP*CP*A)-3'), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Wang, C, Zou, T.T. | | Deposit date: | 2024-09-20 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | DNA methylation activates retron Ec86 filaments for antiphage defense.
Cell Rep, 43, 2024
|
|
3FF9
 
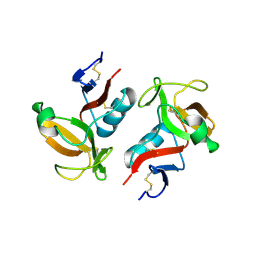 | | Structure of NK cell receptor KLRG1 | | Descriptor: | Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
3FF8
 
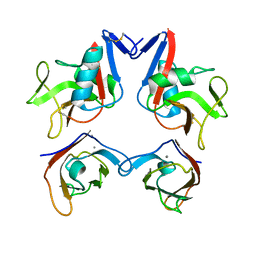 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | Descriptor: | CALCIUM ION, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
6LNA
 
 | | YdiU complex with AMPNPP and Mn2+ | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
3URF
 
 | | Human RANKL/OPG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 11, soluble form, ... | | Authors: | Wang, X.Q, Luan, X.D, Lu, Q.Y. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of Human RANKL Complexed with Its Decoy Receptor Osteoprotegerin
J.Immunol., 189, 2012
|
|
3R22
 
 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesana, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4TVZ
 
 | | Crystal Structure of SCARB2 in Neural Condition (pH7.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor class B member 2, ... | | Authors: | Dang, M.H, Wang, X.X, Rao, Z.H. | | Deposit date: | 2014-06-29 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
4TW0
 
 | | Crystal Structure of SCARB2 in Acidic Condition (pH4.8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dang, M.H, Wang, X.X, Rao, Z.H. | | Deposit date: | 2014-06-29 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.648 Å) | | Cite: | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
3R21
 
 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7MFH
 
 | | Crystal structure of BIO-32546 bound mouse Autotaxin | | Descriptor: | (1R,3S,5S)-8-{(1S)-1-[8-(trifluoromethyl)-7-{[(1s,4R)-4-(trifluoromethyl)cyclohexyl]oxy}naphthalen-2-yl]ethyl}-8-azabicyclo[3.2.1]octane-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chodaparambil, J.V. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent Selective Nonzinc Binding Autotaxin Inhibitor BIO-32546.
Acs Med.Chem.Lett., 12, 2021
|
|
4QR5
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
5IL1
 
 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
