7CI0
 
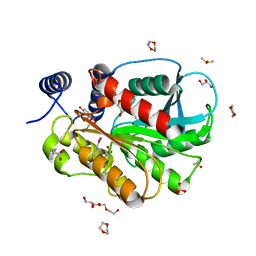 | | Microbial Hormone-sensitive lipase E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
7CIH
 
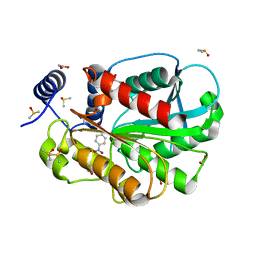 | | Microbial Hormone-sensitive lipase E53 mutant S285G | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
3HLM
 
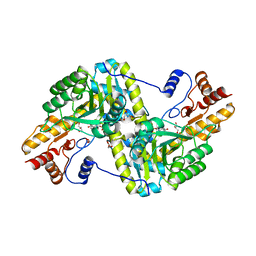 | | Crystal Structure of Mouse Mitochondrial Aspartate Aminotransferase/Kynurenine Aminotransferase IV | | Descriptor: | Aspartate aminotransferase, mitochondrial, GLYCEROL | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2009-05-27 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, expression, and function of kynurenine aminotransferases in human and rodent brains.
Cell.Mol.Life Sci., 67, 2010
|
|
3K40
 
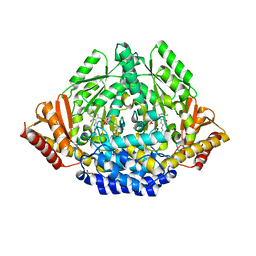 | | Crystal structure of Drosophila 3,4-dihydroxyphenylalanine decarboxylase | | Descriptor: | Aromatic-L-amino-acid decarboxylase, GLYCEROL | | Authors: | Han, Q, Ding, H, Robinson, H, Christensen, B.M, Li, J. | | Deposit date: | 2009-10-05 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and substrate specificity of Drosophila 3,4-dihydroxyphenylalanine decarboxylase
Plos One, 5, 2010
|
|
8K4Y
 
 | | Structure of a triple-helix region of human ReCol 3 from Trautec | | Descriptor: | SULFATE ION, triple-helix region of human ReCol 3 | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human ReCol 3 from Trautec.
To Be Published
|
|
8K4X
 
 | | Structure of a triple-helix region of human Collagen type XVII from Trautec | | Descriptor: | triple-helix region of human Collagen type XVII | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a triple-helix region of human Collagen type XVII from Trautec.
To Be Published
|
|
8K4W
 
 | | Structure of a triple-helix region of human collagen type I from Trautec | | Descriptor: | CHLORIDE ION, triple-helix region of human collagen type I | | Authors: | Chu, Y, Zhai, Y, Fan, X, Li, J, Wang, L, Fu, S, Feng, P, Qian, S. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a triple-helix region of human collagen type I from Trautec.
To Be Published
|
|
6J72
 
 | | Crystal structure of IniA from Mycobacterium smegmatis with GTP bound | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoniazid inducible gene protein IniA, L(+)-TARTARIC ACID, ... | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
2HOB
 
 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | Authors: | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | Deposit date: | 2006-07-14 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
6J73
 
 | | Crystal structure of IniA from Mycobacterium smegmatis | | Descriptor: | Isoniazid inducible gene protein IniA | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
4E11
 
 | | Crystal structure of kynurenine formamidase from Drosophila melanogaster | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2012-03-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster.
Biochem.J., 446, 2012
|
|
4E14
 
 | | Crystal structure of kynurenine formamidase conjugated with phenylmethylsulfonyl fluoride | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, kynurenine formamidase | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2012-03-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster.
Biochem.J., 446, 2012
|
|
3UJL
 
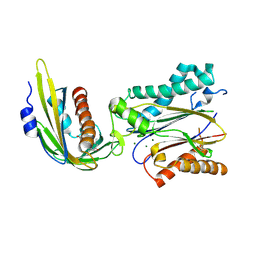 | | Crystal structure of abscisic acid bound PYL2 in complex with type 2C protein phosphatase ABI2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Soon, F.-F, Ng, L.-M, Kovach, A, Tan, M.H.E, Suino-Powell, K.M, He, Y, Xu, Y, Brunzelle, J.S, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Science, 335, 2012
|
|
4E15
 
 | |
2HUF
 
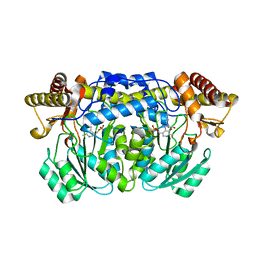 | | Crystal structure of Aedes aegypti alanine glyoxylate aminotransferase | | Descriptor: | 1-BUTANOL, Alanine glyoxylate aminotransferase | | Authors: | Han, Q, Robinson, H, Gao, Y.G, Vogelaar, N, Wilson, S.R, Rizzi, M, Li, J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase.
J.Biol.Chem., 281, 2006
|
|
3N9T
 
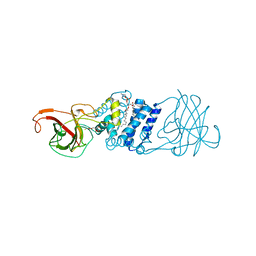 | | Cryatal structure of Hydroxyquinol 1,2-dioxygenase from Pseudomonas putida DLL-E4 | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, CITRATE ANION, FE (III) ION, ... | | Authors: | Liu, W, Shen, W, Fang, P, Li, J, Cui, Z. | | Deposit date: | 2010-05-31 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryatal structure of Hydroxyquinol 1,2-dioxygenase from Pseudomonas putida DLL-E4
To be Published
|
|
4E01
 
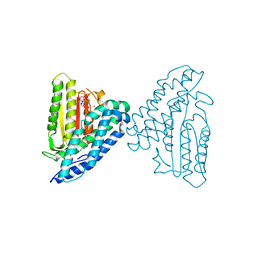 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with AMPPNP | | Descriptor: | 3,6-dichloro-1-benzothiophene-2-carboxylic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
4E00
 
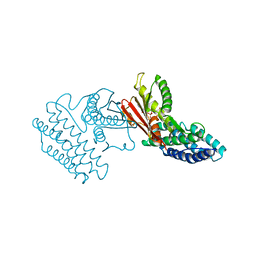 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with ADP | | Descriptor: | 3,6-dichloro-1-benzothiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/3,6-dichlorobenzo[b]thiophene-2-carboxylic acid complex with ADP
To be Published
|
|
4E02
 
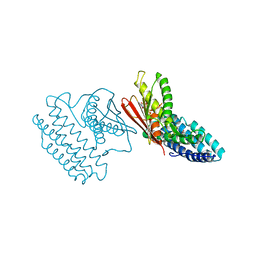 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with AMPPNP | | Descriptor: | (S)-2-chloro-3-phenylpropanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1547 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
8IO3
 
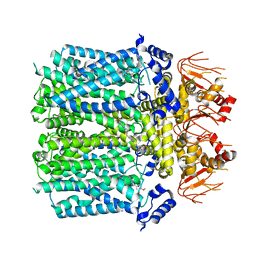 | | Cryo-EM structure of human HCN3 channel with cilobradine | | Descriptor: | 3-[[(3~{S})-1-[2-(3,4-dimethoxyphenyl)ethyl]piperidin-3-yl]methyl]-7,8-dimethoxy-2,5-dihydro-1~{H}-3-benzazepin-4-one, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure of human HCN3 channel with cilobradine
To Be Published
|
|
8JFQ
 
 | | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-back Loop | | Descriptor: | 26mer-DNA | | Authors: | Liu, Y, Li, J, Zhang, Y, Wang, Y, Chen, J, Bian, Y, Xia, Y, Yang, M.H, Zheng, K, Wang, K.B, Kong, L.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-Back Loop.
J.Am.Chem.Soc., 145, 2023
|
|
1OLU
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-15 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
7EGG
 
 | | TFIID lobe B subcomplex | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 4, ... | | Authors: | Chen, X, Wu, Z, Li, J, Zhao, D, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EDX
 
 | | p53-bound TFIID-based core PIC on HDM2 promoter | | Descriptor: | DNA (84-mer), DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Qi, Y, Hou, H, Wang, X, Wu, Z, Li, J, Xu, Y. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
7EGB
 
 | | TFIID-based holo PIC on SCP promoter | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wu, Z, Hou, H, Qi, Y, Wang, X, Li, J, Xu, Y. | | Deposit date: | 2021-03-24 | | Release date: | 2021-05-05 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into preinitiation complex assembly on core promoters.
Science, 372, 2021
|
|
