8BA5
 
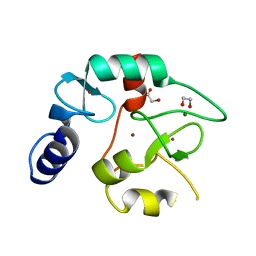 | | Crystal structure of the DNMT3A ADD domain | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, MAGNESIUM ION, ... | | Authors: | Linhard, V, Kunert, S, Wollenhaupt, J, Broehm, A, Schwalbe, H, Jeltsch, A. | | Deposit date: | 2022-10-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The MECP2-TRD domain interacts with the DNMT3A-ADD domain at the H3-tail binding site.
Protein Sci., 32, 2023
|
|
6ELJ
 
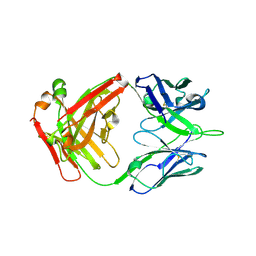 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6ELE
 
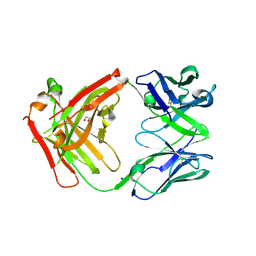 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, fAB heavy chain, ... | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6ELL
 
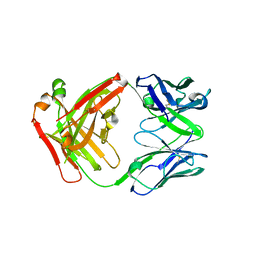 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | fAB heavy chain, fAB light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-09-29 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6EMJ
 
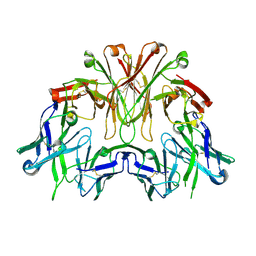 | | FAB Fragment. AbVance: Increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery | | Descriptor: | SODIUM ION, fAB heavy chain, fAb light chain | | Authors: | Benz, J, Weigand, S, Dengl, S, Schlothauer, T, Auer, J, Ehler, A, Kettenberger, H, Lorenz, S, Hirschheydt, T, Georges, G. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AbVance: increasing our knowledge of antibody structural space to enable faster and better decision making in antibody drug discovery
To Be Published
|
|
6GY9
 
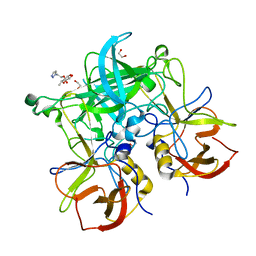 | | Fucose-functionalized precision glycomacromolecules targeting human norovirus capsid protein | | Descriptor: | 1,2-ETHANEDIOL, 2-(1H-1,2,3-triazol-1-yl)ethyl 6-deoxy-alpha-L-galactopyranoside, Capsid protein | | Authors: | Ruoff, K, Kilic, T, Hansman, G.S. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fucose-Functionalized Precision Glycomacromolecules Targeting Human Norovirus Capsid Protein.
Biomacromolecules, 19, 2018
|
|
4YZD
 
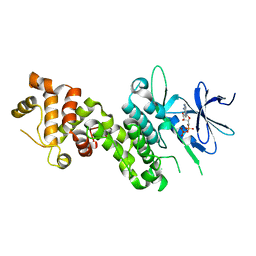 | | Crystal Structure of human phosphorylated IRE1alpha in complex with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
6QCJ
 
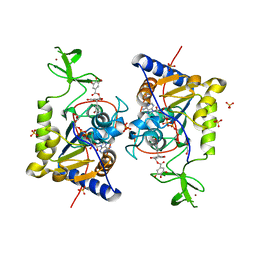 | | Human Sirt6 in complex with ADP-ribose and the inhibitor catechin gallate | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2018-12-28 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives.
Sci Rep, 9, 2019
|
|
4R9M
 
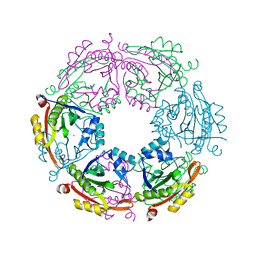 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | Descriptor: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5CNP
 
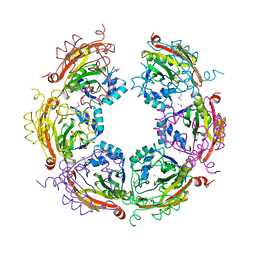 | | X-ray crystal structure of Spermidine n1-acetyltransferase from Vibrio cholerae. | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Osipiuk, J, VOLKART, L, MOY, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
3ZXH
 
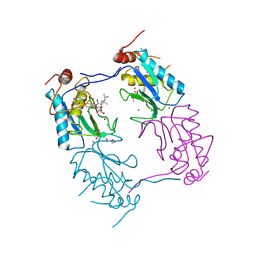 | |
3J07
 
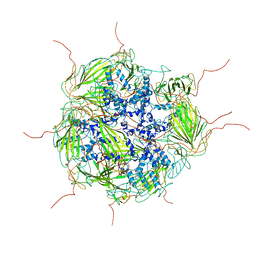 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7KDT
 
 | |
7O00
 
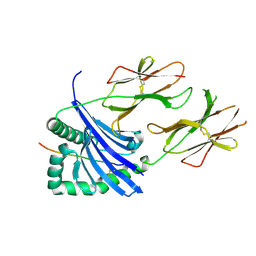 | | Crystal structure of HLA-DR4 in complex with a HSP70 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaperone protein DnaK, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2021-03-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZH
 
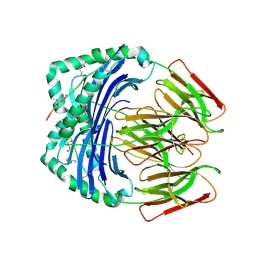 | | Crystal structure of HLA-DR4 in complex with a citrullinated cilp peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.831 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZF
 
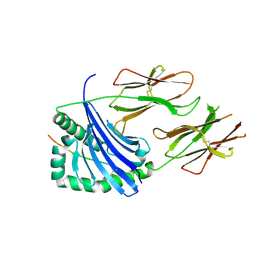 | | Crystal structure of HLA-DR4 in complex with a mutated human collagen type II peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ge, C, Dobritzsch, D, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
7NZE
 
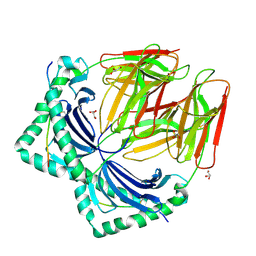 | | Crystal structure of HLA-DR4 in complex with a human collagen type II peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Collagen alpha-1(II) chain, GLYCEROL, ... | | Authors: | Ge, C, Dobritzsch, D, Holmdahl, R. | | Deposit date: | 2021-03-24 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ann Rheum Dis, 81, 2022
|
|
6WZX
 
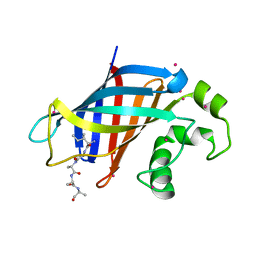 | | GID4 in complex with IGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, ILE-GLY-LEU-TRP-LYS peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8BVI
 
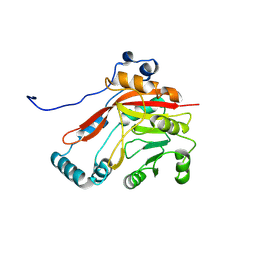 | |
6XUZ
 
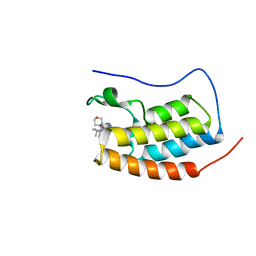 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 4 | | Descriptor: | 6-[1-[(2~{S})-1-methoxypropan-2-yl]-6-[(3~{S})-3-methylmorpholin-4-yl]imidazo[4,5-c]pyridin-2-yl]-3-methyl-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XV3
 
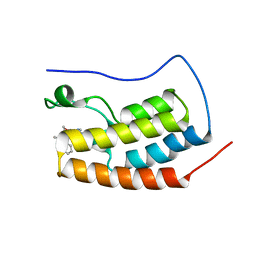 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 3 | | Descriptor: | 3-methyl-6-[6-[(3~{S})-3-methylmorpholin-4-yl]-1-[(1~{S})-1-phenylethyl]imidazo[4,5-c]pyridin-2-yl]-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5MN9
 
 | |
6XVC
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 1 | | Descriptor: | (4~{R})-4-[(1~{R})-1-[7-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)quinolin-5-yl]oxyethyl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.098 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XV7
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 2 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[3,4-bis(fluoranyl)phenyl]methyl]-~{N},3-dimethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6BHG
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
