8HEI
 
 | | Crystal structure of CTSB in complex with E64d | | Descriptor: | Cathepsin B, GLYCEROL, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | Descriptor: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | Descriptor: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HET
 
 | | Crystal structure of CTSL in complex with E64d | | Descriptor: | Procathepsin L, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | Descriptor: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6H
 
 | | Light SFX structure of D.m(6-4)photolyase at 2ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
7AT6
 
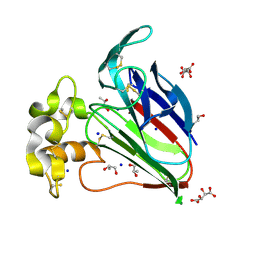 | | Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | L(+)-TARTARIC ACID, R-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Wang, M, Pedrini, B. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
5H2L
 
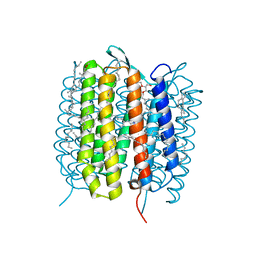 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 5.25 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2N
 
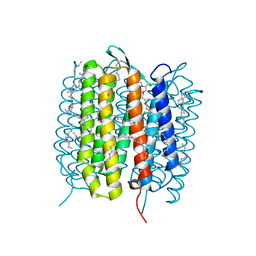 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 95.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2O
 
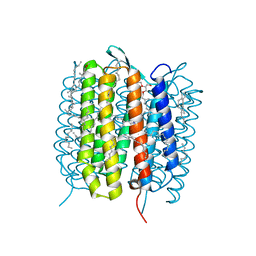 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 250 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2M
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 13.8 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
8S4F
 
 | | Human carbonic anhydrase I covalently bound to AV21-08 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(cyclooctylamino)-4-ethylsulfonyl-2,5,6-tris(fluoranyl)benzenesulfonamide, ... | | Authors: | Manakova, E.N, Grazulis, S, Paketuryte-Latve, V, Vaskevicius, A, Smirnov, A. | | Deposit date: | 2024-02-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Targeted anticancer pre-vinylsulfone covalent inhibitors of carbonic anhydrase IX.
Elife, 13, 2024
|
|
3L0R
 
 | | Crystal Structure of Salivary Cystatin from the Soft Tick Ornithodoros moubata | | Descriptor: | CHLORIDE ION, Cystatin-2, GLYCEROL | | Authors: | Rezacova, P, Brynda, J, Andersen, J.F, Salat, J, Kovarova, Z, Mares, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and functional characterization of an immunomodulatory salivary cystatin from the soft tick Ornithodoros moubata
Biochem.J., 429, 2010
|
|
5H2J
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 290 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2P
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 657 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2H
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 40 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2K
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2I
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 110 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
