6TQF
 
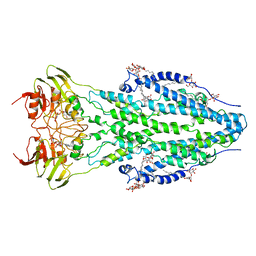 | | The structure of ABC transporter Rv1819c in AMP-PNP bound state | | Descriptor: | ABC transporter ATP-binding protein/permease, DODECYL-BETA-D-MALTOSIDE, MAGNESIUM ION, ... | | Authors: | Rempel, S, Gati, C, Slotboom, D.J, Guskov, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-04-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds.
Nature, 580, 2020
|
|
1KH5
 
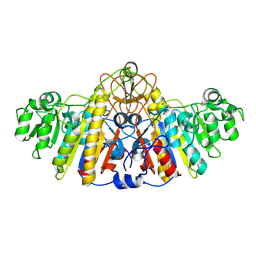 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALKALINE PHOSPHATASE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHJ
 
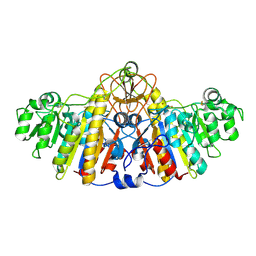 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALUMINUM FLUORIDE, Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KR3
 
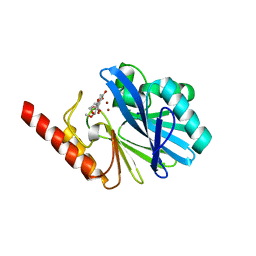 | | Crystal Structure of the Metallo beta-Lactamase from Bacteroides fragilis (CfiA) in Complex with the Tricyclic Inhibitor SB-236050. | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Payne, D.J, Hueso-Rodrguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Cheever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Dez, E, Prez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-beta-lactamases
ANTIMICROB.AGENTS CHEMOTHER., 46, 2002
|
|
4LJ8
 
 | | ClpB NBD2 R621Q from T. thermophilus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1KSE
 
 | | Solution Structure of a quinolone-capped DNA duplex | | Descriptor: | 5'-D(*(5AT)P*GP*CP*GP*CP*A)-3', OXOLINIC ACID | | Authors: | Tuma, J, Connors, W.H, Stitelman, D.H, Richert, C. | | Deposit date: | 2002-01-12 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | On the effect of covalently appended quinolones on termini of DNA duplexes.
J.Am.Chem.Soc., 124, 2002
|
|
4LLP
 
 | | Crystal structure of PDE10A2 with fragment ZT401 | | Descriptor: | 4-amino-2-methylphenol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM4
 
 | | Crystal structure of PDE10A2 with fragment ZT902 | | Descriptor: | NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, quinazolin-4(1H)-one | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-10 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LMW
 
 | |
1KTM
 
 | |
1KV7
 
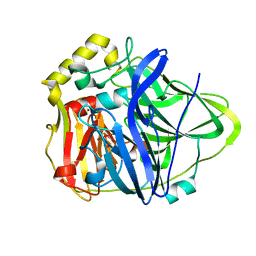 | | Crystal Structure of CueO, a multi-copper oxidase from E. coli involved in copper homeostasis | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, PROBABLE BLUE-COPPER PROTEIN YACK | | Authors: | Roberts, S.A, Weichsel, A, Grass, G, Thakali, K, Hazzard, J.T, Tollin, G, Rensing, C, Montfort, W.R. | | Deposit date: | 2002-01-25 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KV2
 
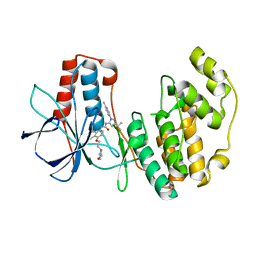 | | Human p38 MAP Kinase in Complex with BIRB 796 | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
4LAY
 
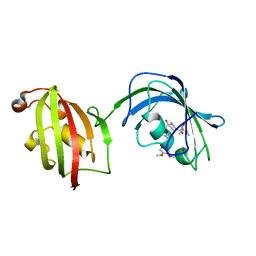 | | Crystal Structure Analysis of FKBP52, Complex with I63 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, {3-[(1R)-3-(3,4-dimethoxyphenyl)-1-({[(2S)-1-(3,3-dimethyl-2-oxopentanoyl)piperidin-2-yl]carbonyl}oxy)propyl]phenoxy}acetic acid | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
1KWI
 
 | | Crystal Structure Analysis of the Cathelicidin Motif of Protegrins | | Descriptor: | Protegrin-3 Precursor | | Authors: | Sanchez, J.F, Hoh, F, Strub, M.P, Aumelas, A, Dumas, C. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the cathelicidin motif of protegrin-3 precursor: structural insights into the activation mechanism of an antimicrobial protein.
Structure, 10, 2002
|
|
4LAV
 
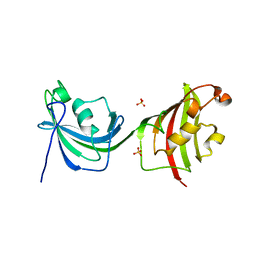 | | Crystal Structure Analysis of FKBP52, Crystal Form II | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, SULFATE ION | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
1KXQ
 
 | | Camelid VHH Domain in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | CALCIUM ION, CHLORIDE ION, alpha-amylase, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
4LKX
 
 | | Humanized antibody 4B12 Fab complexed with a CemX segment | | Descriptor: | CemX segment, Fab fragment heavy chain, Fab fragment light chain | | Authors: | Chu, H.M, Wright, J, Chan, Y.H, Lin, C.J, Chang, T.W, Lim, C. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Two potential therapeutic antibodies bind to a peptide segment of membrane-bound IgE in different conformations.
Nat Commun, 5, 2014
|
|
4LM1
 
 | | Crystal structure of PDE10A2 with fragment ZT450 | | Descriptor: | 5-nitroquinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
2I59
 
 | | Solution structure of RGS10 | | Descriptor: | Regulator of G-protein signaling 10 | | Authors: | Fedorov, O, Higman, V.A, Diehl, A, Leidert, M, Lemak, A, Schmieder, P, Oschkinat, H, Elkins, J, Soundarajan, M, Doyle, D.A, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4LAW
 
 | | Crystal Structure Analysis of FKBP52, Crystal Form III | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
1KV1
 
 | | p38 MAP Kinase in Complex with Inhibitor 1 | | Descriptor: | 1-(5-TERT-BUTYL-2-METHYL-2H-PYRAZOL-3-YL)-3-(4-CHLORO-PHENYL)-UREA, p38 MAP kinase | | Authors: | Pargellis, C, Tong, L, Churchill, L, Cirillo, P.F, Gilmore, T, Graham, A.G, Grob, P.M, Hickey, E.R, Moss, N, Pav, S, Regan, J. | | Deposit date: | 2002-01-23 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site.
Nat.Struct.Biol., 9, 2002
|
|
4LLX
 
 | | Crystal structure of PDE10A2 with fragment ZT434 | | Descriptor: | 4,6-dimethylpyrimidin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LJ4
 
 | | ClpB NBD2 from T. thermophilus, nucleotide-free | | Descriptor: | Chaperone protein ClpB, PHOSPHATE ION | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2HQI
 
 | | NMR SOLUTION STRUCTURE OF THE OXIDIZED FORM OF MERP, 14 STRUCTURES | | Descriptor: | MERCURIC TRANSPORT PROTEIN | | Authors: | Qian, H, Sahlman, L, Eriksson, P.O, Hambreus, C, Edlund, U, Sethson, I. | | Deposit date: | 1998-03-31 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the oxidized form of MerP, a mercuric ion binding protein involved in bacterial mercuric ion resistance.
Biochemistry, 37, 1998
|
|
4LDF
 
 | | Crystal Structure of CpBRD2 from cryptosporidium, cgd3_3190 | | Descriptor: | GCN5 like acetylase + bromodomain, GLYCEROL, UNKNOWN ATOM OR ION | | Authors: | Wernimont, A.K, Loppnau, P, Fonseca, M, Knapp, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Mottaghi, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of CpBRD2 from cryptosporidium, cgd3_3190
TO BE PUBLISHED
|
|
