2YNH
 
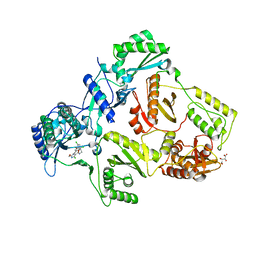 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK500 | | Descriptor: | 4-chloranyl-N-[[4-chloranyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-2-(hydroxymethyl)-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
3BF3
 
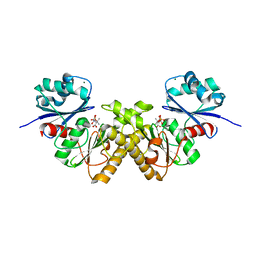 | | Type III pantothenate kinase from Thermotoga maritima complexed with product phosphopantothenate | | Descriptor: | MAGNESIUM ION, N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanine, Type III pantothenate kinase | | Authors: | Yang, K, Huerta, C, Strauss, E, Zhang, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for substrate binding and the catalytic mechanism of type III pantothenate kinase.
Biochemistry, 47, 2008
|
|
8JYU
 
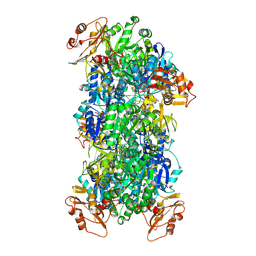 | | Acyl-ACP Synthetase structure bound to Decanoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, DECANOIC ACID, ... | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | An inhibitory mechanism of AasS, an exogenous fatty acid scavenger: Implications for re-sensitization of FAS II antimicrobials.
Plos Pathog., 20, 2024
|
|
8JYL
 
 | | Acyl-ACP Synthetase structure bound to C10-AMS | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-decanoylsulfamate | | Authors: | Huang, H, Chang, S, Huang, M, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-07-03 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | An inhibitory mechanism of AasS, an exogenous fatty acid scavenger: Implications for re-sensitization of FAS II antimicrobials.
Plos Pathog., 20, 2024
|
|
3BF1
 
 | | Type III pantothenate kinase from Thermotoga maritima complexed with pantothenate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PANTOTHENOIC ACID, Type III pantothenate kinase | | Authors: | Yang, K, Huerta, C, Strauss, E, Zhang, H. | | Deposit date: | 2007-11-20 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for substrate binding and the catalytic mechanism of type III pantothenate kinase.
Biochemistry, 47, 2008
|
|
7CJF
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Guo, Y, Li, X, Zhang, G, Fu, D, Schweizer, L, Zhang, H, Rao, Z. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody with extensive Spike binding coverage and modified for optimal therapeutic outcomes.
Nat Commun, 12, 2021
|
|
8HEF
 
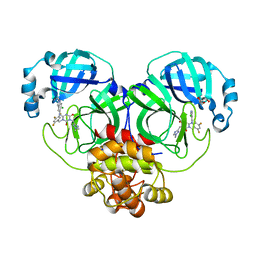 | | The Crystal structure of deuterated S-217622 (Ensitrelvir) bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Synthesis of deuterated S-217622 (Ensitrelvir) with antiviral activity against coronaviruses including SARS-CoV-2.
Antiviral Res., 213, 2023
|
|
5WSZ
 
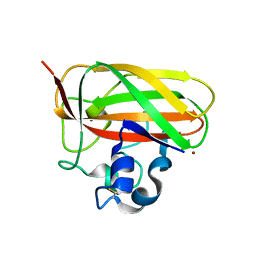 | | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis | | Descriptor: | COPPER (II) ION, LpmO10A | | Authors: | Zhao, Y, Zhang, H, Yin, H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis
To Be Published
|
|
8IKJ
 
 | | Cryo-EM structure of the inactive CD97 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor E5,Soluble cytochrome b562,Adhesion G protein-coupled receptor E5 subunit beta | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P, Guo, J, Qin, J, Shen, D, Ji, S, Zang, S, Zhang, H, Wang, W, Shen, Q, Sun, P, Zhang, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
5V7U
 
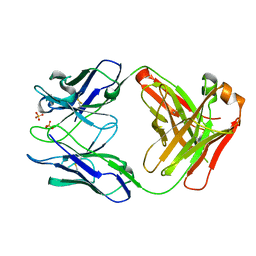 | |
8ZLU
 
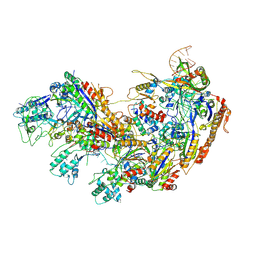 | | Cryo-EM strcuture of Cas5-HNH Cascade,Conf1 | | Descriptor: | CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, CRISPR-associated endoribonuclease Cse3, ... | | Authors: | Liu, Y.N, Wang, L, Zhang, H, Zhu, H. | | Deposit date: | 2024-05-21 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural basis for RNA-guided DNA degradation by Cas5-HNH/Cascade complex.
Nat Commun, 16, 2025
|
|
8ZP7
 
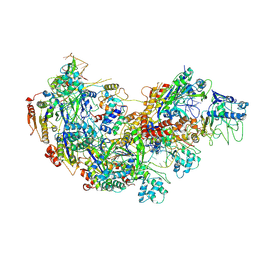 | | Cryo-EM structure of Cas5-HNH Cascade bound with sDNA, Conf1 | | Descriptor: | CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, CRISPR-associated endoribonuclease Cse3, ... | | Authors: | Liu, Y.N, Wang, L, Zhang, H, Zhu, H. | | Deposit date: | 2024-05-29 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for RNA-guided DNA degradation by Cas5-HNH/Cascade complex.
Nat Commun, 16, 2025
|
|
8ZP9
 
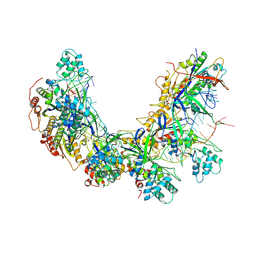 | | Cryo-EM structure of Cas5-HNH Cascade bound with sDNA, Conf2 | | Descriptor: | CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, DNA (60-MER), ... | | Authors: | Liu, Y.N, Wang, L, Zhang, H, Zhu, H. | | Deposit date: | 2024-05-29 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for RNA-guided DNA degradation by Cas5-HNH/Cascade complex.
Nat Commun, 16, 2025
|
|
8ZM3
 
 | | Cryo-EM strcuture of Cas5-HNH Cascade,apo-Conf2 | | Descriptor: | CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, CRISPR-associated endoribonuclease Cse3, ... | | Authors: | Liu, Y.N, Wang, L, Zhang, H, Zhu, H. | | Deposit date: | 2024-05-22 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for RNA-guided DNA degradation by Cas5-HNH/Cascade complex.
Nat Commun, 16, 2025
|
|
8ZOL
 
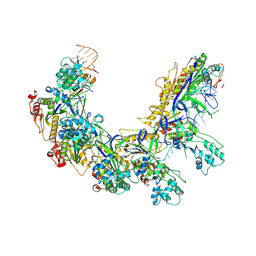 | | Cryo-EM strcuture of Cas5-HNH Cascade,Conf3 | | Descriptor: | CRISPR system Cascade subunit CasC, CRISPR system Cascade subunit CasD, CRISPR-associated endoribonuclease Cse3, ... | | Authors: | Liu, Y.N, Wang, L, Zhang, H, Zhu, H. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural basis for RNA-guided DNA degradation by Cas5-HNH/Cascade complex.
Nat Commun, 16, 2025
|
|
8UCD
 
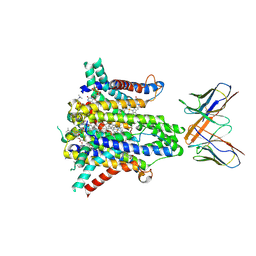 | | Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, AMG 509 anti-STEAP1 Fab, heavy chain, ... | | Authors: | Li, F, Bailis, J.M, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AMG 509 (Xaluritamig), an Anti-STEAP1 XmAb 2+1 T-cell Redirecting Immune Therapy with Avidity-Dependent Activity against Prostate Cancer.
Cancer Discov, 14, 2024
|
|
5V7R
 
 | |
8W9X
 
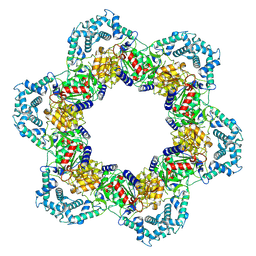 | |
8W9L
 
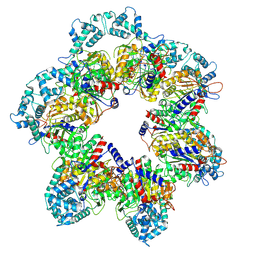 | |
2MWN
 
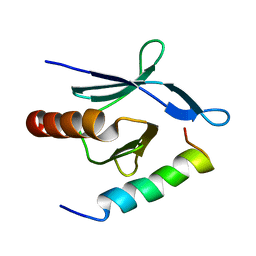 | | Talin-F3 / RIAM N-terminal Peptide complex | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, Talin-1 | | Authors: | Yang, J, Zhu, L, Zhang, H, Hirbawi, J, Fukuda, K, Dwivedi, P, Liu, J, Byzova, T, Plow, E.F, Wu, J, Qin, J. | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational activation of talin by RIAM triggers integrin-mediated cell adhesion.
Nat Commun, 5, 2014
|
|
2MPX
 
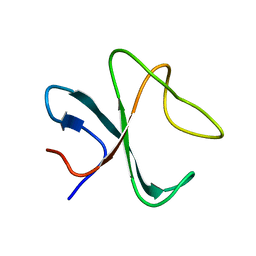 | |
2DHS
 
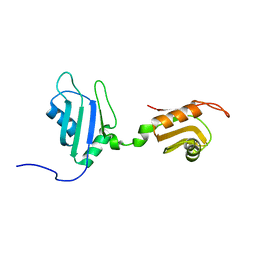 | | Solution Structure of Nucleic Acid Binding Protein CUGBP1ab and its Binding Study with DNA and RNA | | Descriptor: | CUG triplet repeat RNA-binding protein 1 | | Authors: | Xia, Y.L, Jun, K.Y, Zhu, Q, Han, X.G, Zhang, H, Timchenko, L, Swanson, M, Gao, X.L. | | Deposit date: | 2006-03-25 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Nucleic Acid Binding Protein CUGBP1ab and its Binding Study with DNA and RNA
To be Published
|
|
2E74
 
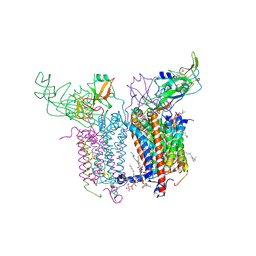 | | Crystal Structure of the Cytochrome b6f Complex from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, Apocytochrome f, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
2E75
 
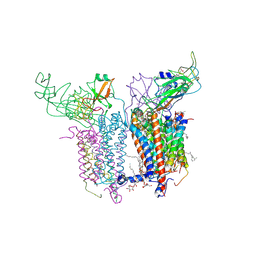 | | Crystal Structure of the Cytochrome b6f Complex with 2-nonyl-4-hydroxyquinoline N-oxide (NQNO) from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2-NONYL-4-HYDROXYQUINOLINE N-OXIDE, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
