8HDJ
 
 | |
8HER
 
 | |
8HEP
 
 | |
8HEQ
 
 | |
1VIO
 
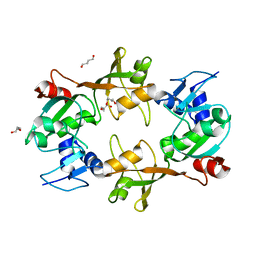 | | Crystal structure of pseudouridylate synthase | | Descriptor: | 1,4-BUTANEDIOL, Ribosomal small subunit pseudouridine synthase A | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the pseudouridine synthase RsuA from Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VJE
 
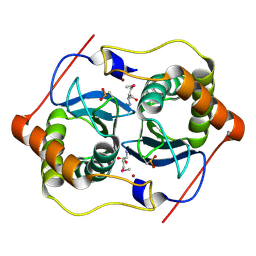 | |
6RZX
 
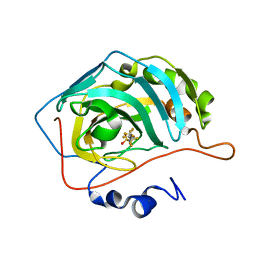 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
7CJR
 
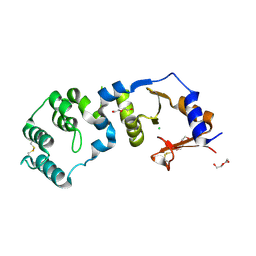 | | Crystal structure of a periplasmic sensor domain of histidine kinase VbrK | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Histidine kinase | | Authors: | Goh, B.C, Chua, Y.K, Qian, X, Savko, M, Lescar, J. | | Deposit date: | 2020-07-12 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the periplasmic sensor domain of histidine kinase VbrK suggests indirect sensing of beta-lactam antibiotics.
J.Struct.Biol., 212, 2020
|
|
6K4F
 
 | | SiaC of Pseudomonas aeruginosa | | Descriptor: | DUF1987 domain-containing protein | | Authors: | Lin, J.Q, Lescar, J. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | The SiaABC threonine phosphorylation pathway controls biofilm formation in response to carbon availability in Pseudomonas aeruginosa.
Plos One, 15, 2020
|
|
6K4E
 
 | | SiaA-PP2C domain of Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, Putative serine phosphatase | | Authors: | Lin, J.Q, Lescar, J. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | The SiaABC threonine phosphorylation pathway controls biofilm formation in response to carbon availability in Pseudomonas aeruginosa.
Plos One, 15, 2020
|
|
4GMI
 
 | | BACE-1 in complex with HEA-type macrocyclic inhibitor, MV078571 | | Descriptor: | (4S,8E)-4-[(1R)-2-{[2-(5-tert-butyl-1,3-oxazol-2-yl)propan-2-yl]amino}-1-hydroxyethyl]-16-methyl-6-oxa-3-azabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraene-2,13-dione, ACETATE ION, Beta-secretase 1, ... | | Authors: | Lindberg, J.D, Derbyshire, D. | | Deposit date: | 2012-08-16 | | Release date: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of novel BACE-1 inhibitors
To be Published
|
|
3ICX
 
 | | Crystal structure of Sulfolobus solfataricus Nop5 (135-380) | | Descriptor: | Pre mRNA splicing protein, SULFATE ION | | Authors: | Ye, K. | | Deposit date: | 2009-07-19 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural organization of box C/D RNA-guided RNA methyltransferase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5XM9
 
 | |
5XMA
 
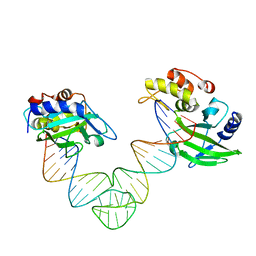 | |
5VJH
 
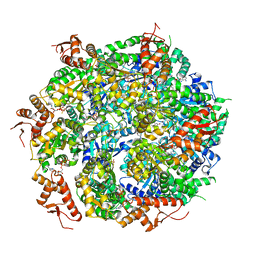 | | Closed State CryoEM Reconstruction of Hsp104:ATPyS and FITC casein | | Descriptor: | FITC casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
8TJF
 
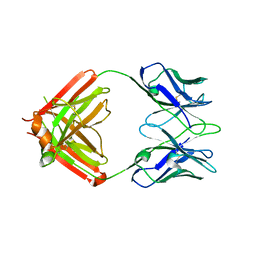 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | Fab Lambda light chain, IgG1 Fab heavy chain | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y, Chiang, C. | | Deposit date: | 2023-07-21 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
8TI4
 
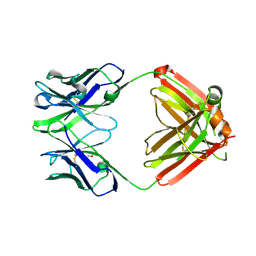 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | GLYCEROL, IgG1 Fab heavy chain, mutated to promote correct pairing, ... | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
5VY8
 
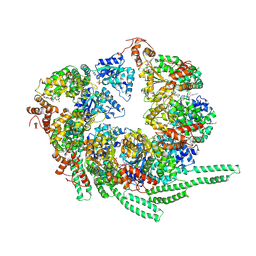 | | S. cerevisiae Hsp104-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 104 | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5XM8
 
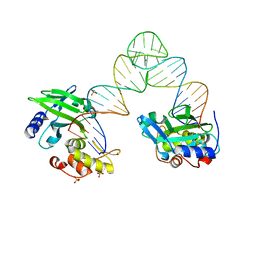 | |
5VYA
 
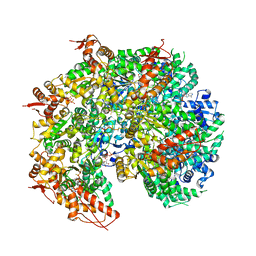 | | S. cerevisiae Hsp104:casein complex, Extended Conformation | | Descriptor: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
9EST
 
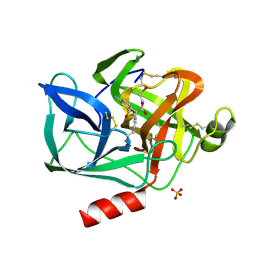 | | STRUCTURAL STUDY OF PORCINE PANCREATIC ELASTASE COMPLEXED WITH 7-AMINO-3-(2-BROMOETHOXY)-4-CHLOROISOCOUMARIN AS A NONREACTIVATABLE DOUBLY COVALENT ENZYME-INHIBITOR COMPLEX | | Descriptor: | (2-BROMOETHYL)(2-'FORMYL-4'-AMINOPHENYL) ACETATE, CALCIUM ION, PORCINE PANCREATIC ELASTASE, ... | | Authors: | Radhakrishnan, R, Powers, J.C, Meyer Jr, E.F. | | Deposit date: | 1991-01-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural study of porcine pancreatic elastase complexed with 7-amino-3-(2-bromoethoxy)-4-chloroisocoumarin as a nonreactivatable doubly covalent enzyme-inhibitor complex.
Biochemistry, 30, 1991
|
|
3ID5
 
 | |
3MVM
 
 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7V | | Descriptor: | 4-{[5-(isoxazol-3-ylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38? Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
3MVL
 
 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7K | | Descriptor: | 4-{[5-(cyclopropylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38 Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
3ID6
 
 | |
