6MDU
 
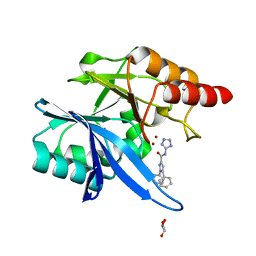 | | Crystal structure of NDM-1 with compound 7 | | Descriptor: | 1,5-diphenyl-N-(1H-tetrazol-5-yl)-1H-pyrazole-3-carboxamide, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
8E2B
 
 | | N-terminal domain of S. aureus GpsB | | Descriptor: | Cell cycle protein GpsB, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB.
Elife, 13, 2024
|
|
8E2C
 
 | |
2JWS
 
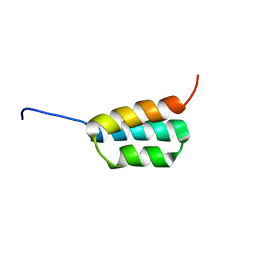 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | Descriptor: | Ga88 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2MH8
 
 | | GA-79-MBP cs-rosetta structures | | Descriptor: | GA-79-MBP, maltose binding protein | | Authors: | He, Y, Chen, Y, Porter, L, Bryan, P, Orban, J. | | Deposit date: | 2013-11-19 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Subdomain interactions foster the design of two protein pairs with 80% sequence identity but different folds.
Biophys.J., 108, 2015
|
|
3C5W
 
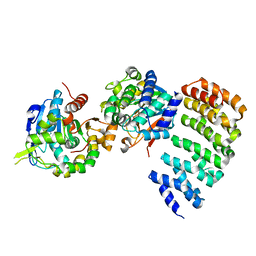 | | Complex between PP2A-specific methylesterase PME-1 and PP2A core enzyme | | Descriptor: | PP2A A subunit, PP2A C subunit, PP2A-specific methylesterase PME-1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
2LHG
 
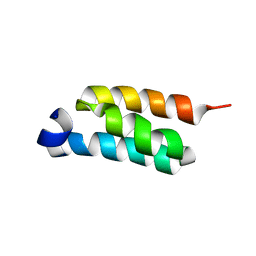 | | GB98-T25I solution structure | | Descriptor: | GB98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2JWU
 
 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | Descriptor: | Gb88 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2007-10-25 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2KDL
 
 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | designed protein | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3C96
 
 | | Crystal structure of the flavin-containing monooxygenase phzS from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR240 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-15 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the flavin-containing monooxygenase phzS from Pseudomonas aeruginosa.
To be Published
|
|
3C5V
 
 | | PP2A-specific methylesterase apo form (PME) | | Descriptor: | Protein phosphatase methylesterase 1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
2LHE
 
 | | Gb98-T25I,L20A | | Descriptor: | Gb98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LHD
 
 | | GB98 solution structure | | Descriptor: | GB98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LU1
 
 | | pfsub2 solution NMR structure | | Descriptor: | Subtilase | | Authors: | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | Deposit date: | 2012-06-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
4ZOU
 
 | |
5A9Y
 
 | | Structure of ppGpp BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9X
 
 | | Structure of GDP bound BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4N4Y
 
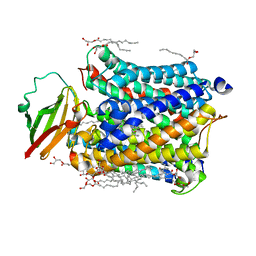 | |
5AA0
 
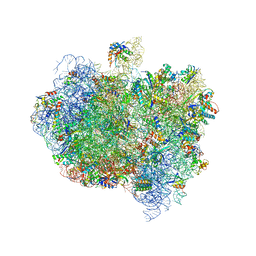 | | Complex of Thermous thermophilus ribosome (A-and P-site tRNA) bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9W
 
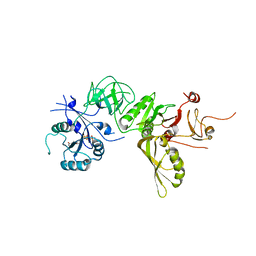 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1RYU
 
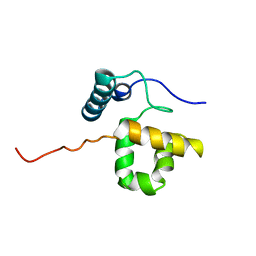 | | Solution Structure of the SWI1 ARID | | Descriptor: | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1 | | Authors: | Kim, S, Zhang, Z, Upchurch, S, Isern, N, Chen, Y. | | Deposit date: | 2003-12-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition.
J.Biol.Chem., 279, 2004
|
|
1R78
 
 | | CDK2 complex with a 4-alkynyl oxindole inhibitor | | Descriptor: | 4-((3R,4S,5R)-4-AMINO-3,5-DIHYDROXY-HEX-1-YNYL)-5-FLUORO-3-[1-(3-METHOXY-1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Luk, K.-C, Simcox, M.E, Schutt, A, Rowan, K, Thompson, T, Chen, Y, Kammlott, U, DePinto, W, Dunten, P, Dermatakis, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new series of potent oxindole inhibitors of CDK2
Bioorg.Med.Chem.Lett., 14, 2004
|
|
5A9Z
 
 | | Complex of Thermous thermophilus ribosome bound to BipA-GDPCP | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Chen, Y, Ahmed, T, Tan, J, Ero, R, Bhushan, S, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9V
 
 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1SQ4
 
 | | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14 | | Descriptor: | Glyoxylate-induced Protein, THIOCYANATE ION | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Glyoxylate Induced Protein from Pseudomonas aeruginosa, Northeast Structural Genomics Target PaR14
To be Published
|
|
