4ZSE
 
 | |
9BPF
 
 | |
1QT1
 
 | |
9BOO
 
 | | Crystal structure of MERS-CoV Nsp5 in complex with PF-07817883 | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (Mpro) of SARS-CoV-2 and MERS-CoV
Jacs Au, 2024
|
|
6RGV
 
 | |
8JIL
 
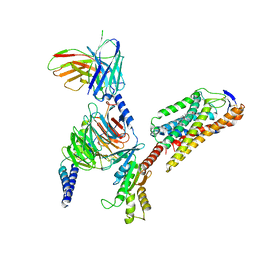 | | Cryo-EM structure of niacin bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JII
 
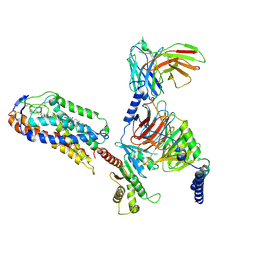 | | Cryo-EM structure of compound 9n and niacin bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JHY
 
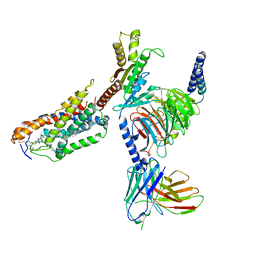 | | Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIM
 
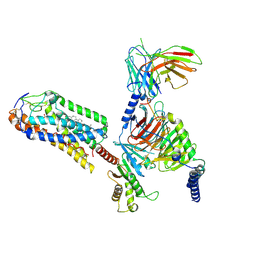 | | Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
5YU9
 
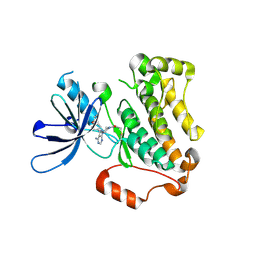 | | Crystal structure of EGFR 696-1022 T790M in complex with Ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ibrutinib targets mutant-EGFR kinase with a distinct binding conformation.
Oncotarget, 7, 2016
|
|
6C0S
 
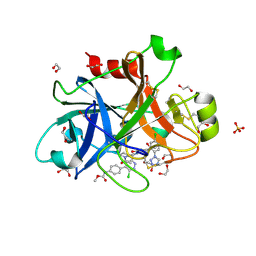 | |
7MXD
 
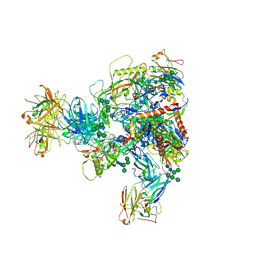 | |
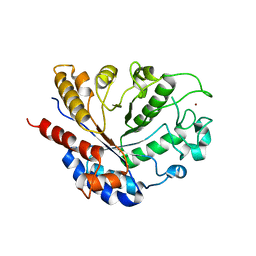
8BAL
 
 | |
8BDP
 
 | |
8BBL
 
 | | SGL a GH20 family sulfoglycosidase | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Dong, M.D, Roth, C.R, Jin, Y.J. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 2022
|
|
1CLK
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES DIASTATICUS NO.7 STRAIN M1033 XYLOSE ISOMERASE AT 1.9 A RESOLUTION WITH PSEUDO-I222 SPACE GROUP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Niu, L, Teng, M, Zhu, X, Gong, W. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of xylose isomerase from Streptomyces diastaticus no. 7 strain M1033 at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7N28
 
 | |
8HJ5
 
 | | Cryo-EM structure of Gq-coupled MRGPRX1 bound with Compound-16 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Gan, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2022-11-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of agonist-induced activation of the human itch receptor MRGPRX1.
Plos Biol., 21, 2023
|
|
8H1R
 
 | | Crystal structure of LptDE-YifL complex | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Lipoprotein sorting to the cell surface via a crosstalk between the Lpt and Lol pathways during outer membrane biogenesis
To Be Published
|
|
8H1S
 
 | | Crystal structure of apo-LptDE complex | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Lipoprotein sorting to the cell surface via a crosstalk between the Lpt and Lol pathways during outer membrane biogenesis
To Be Published
|
|
5W0C
 
 | | Cytochrome P450 (CYP) 2C9 TCA007 Inhibitor Complex | | Descriptor: | Cytochrome P450 2C9, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Johnson, E.F, Hsu, M.-H. | | Deposit date: | 2017-05-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Determinants of the Inhibition of DprE1 and CYP2C9 by Antitubercular Thiophenes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
