1I87
 
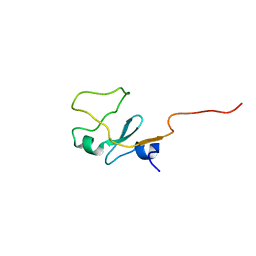 | | SOLUTION STRUCTURE OF THE WATER-SOLUBLE FRAGMENT OF RAT HEPATIC APOCYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 | | Authors: | Falzone, C.J, Wang, Y, Vu, B.C, Scott, N.L, Bhattacharya, S, Lecomte, J.T. | | Deposit date: | 2001-03-12 | | Release date: | 2001-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic perturbations induced by heme binding in cytochrome b5.
Biochemistry, 40, 2001
|
|
1KGW
 
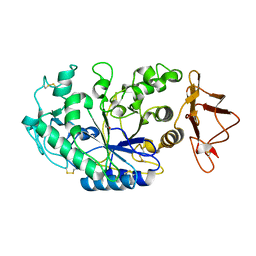 | | THREE DIMENSIONAL STRUCTURE ANALYSIS OF THE R337Q VARIANT OF HUMAN PANCREATIC ALPHA-MYLASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, PANCREATIC, ... | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
1KB3
 
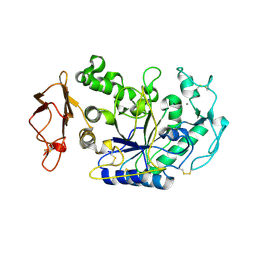 | | Three Dimensional Structure Analysis of the R195A Variant of Human Pancreatic Alpha Amylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, PANCREATIC, ... | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-05 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
1KGX
 
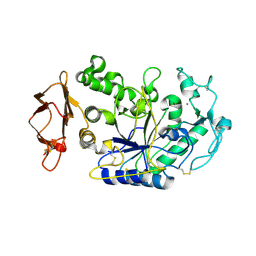 | | Three Dimensional Structure Analysis of the R195Q Variant of Human Pancreatic Alpha Amylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-AMYLASE, PANCREATIC, ... | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
1KGU
 
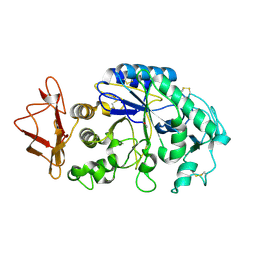 | | THREE DIMENSIONAL STRUCTURE ANALYSIS OF THE R337A VARIANT OF HUMAN PANCREATIC ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION | | Authors: | Numao, S, Maurus, R, Sidhu, G, Wang, Y, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of the chloride ion in the mechanism of human pancreatic alpha-amylase.
Biochemistry, 41, 2002
|
|
5UY8
 
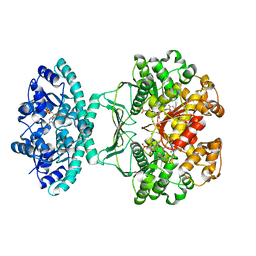 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | 5-[(5S)-5-ethyl-5-methyl-6-oxo-1,4,5,6-tetrahydropyridin-3-yl]-N-(6-fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)thiophene-2-sulfonamide, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, ... | | Authors: | Wang, J, Wang, Y, Fales, K.R, Atwell, S, Clawson, D. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
5UZ0
 
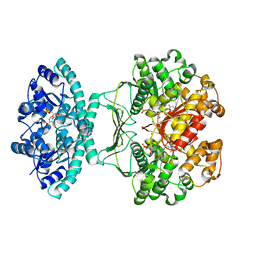 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, MAGNESIUM ION, ... | | Authors: | Atwell, S, Wang, Y, Fales, K.R, Clawson, D, Wang, J. | | Deposit date: | 2017-02-24 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
6K3B
 
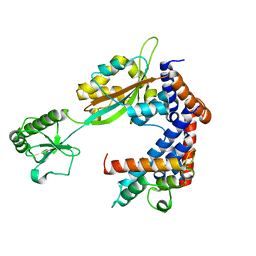 | | Crystal structure of Lpg2147-Lpg2149 complex | | Descriptor: | Lpg2147, Uncharacterized protein | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-05-17 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
2PL5
 
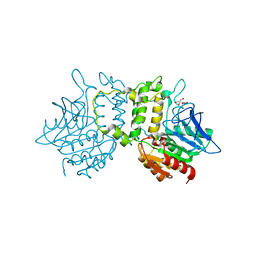 | | Crystal Structure of Homoserine O-acetyltransferase from Leptospira interrogans | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Liu, L, Wang, M, Wang, Y, Wei, Z, Xu, H, Gong, W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of homoserine O-acetyltransferase from Leptospira interrogans
Biochem.Biophys.Res.Commun., 363, 2007
|
|
6KFP
 
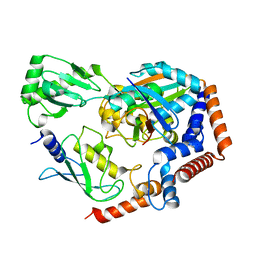 | | Crystal structure of MavC ternary complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
2F96
 
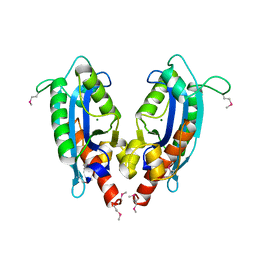 | | 2.1 A crystal structure of Pseudomonas aeruginosa rnase T (Ribonuclease T) | | Descriptor: | MAGNESIUM ION, Ribonuclease T | | Authors: | Zheng, H, Chruszcz, M, Cymborowski, M, Wang, Y, Gorodichtchenskaia, E, Skarina, T, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of RNase T, an Exoribonuclease Involved in tRNA Maturation and End Turnover.
Structure, 15, 2007
|
|
3PMR
 
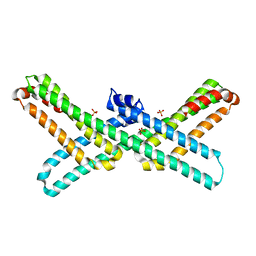 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
6MQC
 
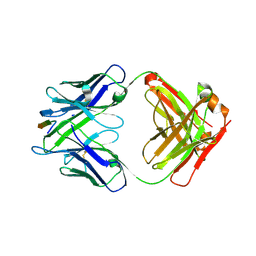 | |
4IG4
 
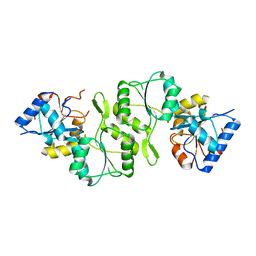 | | Crystal structure of single mutant thermostable NPPase (N86S) from Geobacillus stearothermophilus | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Wang, F, Huang, J, Qiu, R, Yang, Z, Wang, Y, Gong, W, Ji, C. | | Deposit date: | 2012-12-16 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of thermostable NPPase from Geobacillus stearothermophilus
To be Published
|
|
6N16
 
 | |
6MQE
 
 | |
6MQS
 
 | |
6CPT
 
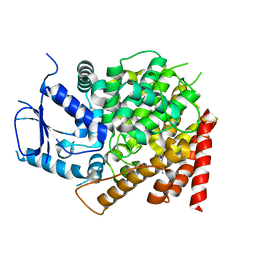 | | crystal structure of yeast caPDE2 in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, Phosphodiesterase, ... | | Authors: | Ke, h, Wang, Y. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Candida albicans Phosphodiesterase 2 and Implications for Its Biological Functions.
Biochemistry, 57, 2018
|
|
6MQM
 
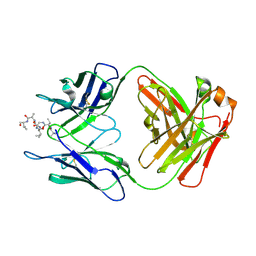 | |
6MQR
 
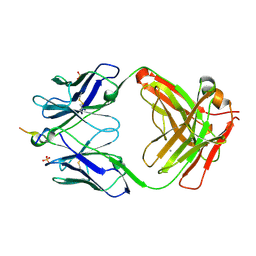 | | Vaccine-elicited NHP FP-targeting neutralizing antibody 0PV-a.01 in complex with FP (residue 512-519) | | Descriptor: | CALCIUM ION, HIV fusion peptide residue 512-519, SULFATE ION, ... | | Authors: | Xu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6CPU
 
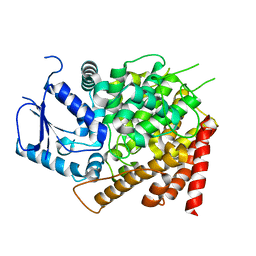 | | Crystal structure of yeast caPDE2 | | Descriptor: | MAGNESIUM ION, Phosphodiesterase, ZINC ION | | Authors: | Ke, H, Wang, y. | | Deposit date: | 2018-03-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Candida albicans Phosphodiesterase 2 and Implications for Its Biological Functions.
Biochemistry, 57, 2018
|
|
4IFT
 
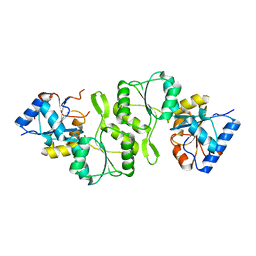 | | Crystal structure of double mutant thermostable NPPase from Geobacillus stearothermophilus | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Huang, J, Wang, F, Qiu, R, Wang, Y, Ji, C. | | Deposit date: | 2012-12-15 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of thermostable NPPase from Geobacillus stearothermophilus
To be Published
|
|
1RE5
 
 | | Crystal structure of 3-carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-carboxy-cis,cis-muconate cycloisomerase, CITRIC ACID | | Authors: | Yang, J, Wang, Y, Woolridge, E.M, Petsko, G.A, Kozarich, J.W, Ringe, D. | | Deposit date: | 2003-11-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of 3-Carboxy-cis,cis-muconate Lactonizing Enzyme from Pseudomonas putida, a Fumarase Class II Type Cycloisomerase: Enzyme Evolution in Parallel Pathways.
Biochemistry, 43, 2004
|
|
1QJL
 
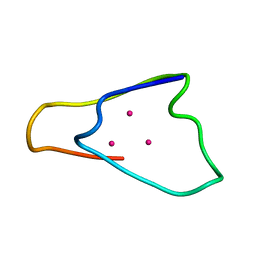 | | METALLOTHIONEIN MTA FROM SEA URCHIN (BETA DOMAIN) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
1QJK
 
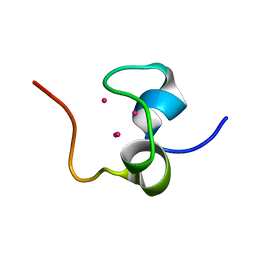 | | Metallothionein MTA from sea urchin (alpha domain) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
