8EXE
 
 | |
8EXF
 
 | |
8FJZ
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(3R,5S)-3-Amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP3
 
 | | PKCeta kinase domain in complex with compound 11 | | Descriptor: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Protein kinase C eta type | | Authors: | Johnson, E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP1
 
 | | PKCeta kinase domain in complex with compound 2 | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | Authors: | Johnson, E. | | Deposit date: | 2023-01-03 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FH4
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | Descriptor: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-13 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | Descriptor: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | McTigue, M, Johnson, E, Cronin, C. | | Deposit date: | 2022-12-21 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8EXI
 
 | |
8EXJ
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain in complex with a JAK1 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK1 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EXN
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with TYK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-receptor tyrosine-protein kinase TYK2 activation loop peptide, PHOSPHATE ION, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8F88
 
 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with monophosphorylated JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EYA
 
 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with a JAK2 activation loop phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EXM
 
 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with a JAK3 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK3 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EYB
 
 | | Crystal structure of PTP1B D181A/Q262A/C215A phosphatase domain with JAK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein kinase JAK2 activation loop phosphopeptide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EXK
 
 | |
9J11
 
 | | Structure of mEos3.2 in the green fluorescent state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Green to red photoconvertible GFP-like protein EosFP | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-03 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9J0R
 
 | | Structure of pcStar in the green fluorescent state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Green to red photoconvertible GFP-like protein EosFP | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9J0Q
 
 | | Structure of mEos3.2 in the green fluorescent state | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Zheng, S.P, Shi, X.R. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural basis for the fast maturation of pcStar, a photoconvertible fluorescent protein.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5GN8
 
 | | Structure of a 48-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | CALCIUM ION, Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Zhao, G, Mikami, B. | | Deposit date: | 2016-07-19 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | "Silent" Amino Acid Residues at Key Subunit Interfaces Regulate the Geometry of Protein Nanocages
ACS Nano, 10, 2016
|
|
9IVP
 
 | | 24-mer DARPin-apoferritin scaffold in complex with the maltose binding protein | | Descriptor: | DARPin,Ferritin heavy chain, N-terminally processed, Maltodextrin-binding protein | | Authors: | Lu, X, Yan, M, Zhang, H.M, Hao, Q. | | Deposit date: | 2024-07-24 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A large, general and modular DARPin-apoferritin scaffold enables the visualization of small proteins by cryo-EM.
Iucrj, 12, 2025
|
|
9BFK
 
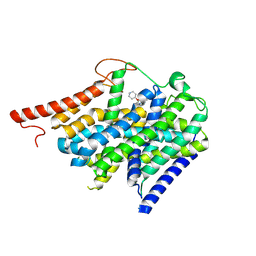 | | Cryo-EM structure of human CHT1 in the ML352 bound state | | Descriptor: | 4-methoxy-3-[(1-methylpiperidin-4-yl)oxy]-N-{[3-(propan-2-yl)-1,2-oxazol-5-yl]methyl}benzamide, High affinity choline transporter 1 | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2024-04-18 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural mechanisms of human sodium-coupled high-affinity choline transporter CHT1.
Cell Discov, 10, 2024
|
|
9BFJ
 
 | | Cryo-EM structure of human CHT1 in the choline bound state | | Descriptor: | CHLORIDE ION, CHOLINE ION, High affinity choline transporter 1, ... | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2024-04-17 | | Release date: | 2024-11-06 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural mechanisms of human sodium-coupled high-affinity choline transporter CHT1.
Cell Discov, 10, 2024
|
|
9BFI
 
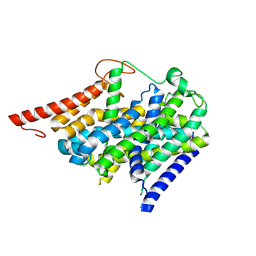 | |
9BIM
 
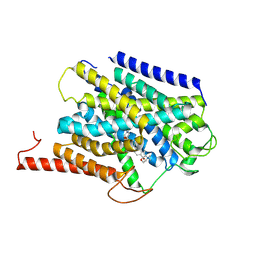 | | Cryo-EM structure of human CHT1 in the HC-3 bound outward-facing state | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), High affinity choline transporter 1 | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-06 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural mechanisms of human sodium-coupled high-affinity choline transporter CHT1.
Cell Discov, 10, 2024
|
|
3PS6
 
 | |
