2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | Descriptor: | Effector protein hopAB3 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | Descriptor: | Effector protein hopAB1 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3H35
 
 | | Structure of the uncharacterized protein ABO_0056 from the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2. | | Descriptor: | 1,2-ETHANEDIOL, S,R MESO-TARTARIC ACID, uncharacterized protein ABO_0056 | | Authors: | Cuff, M.E, Evdokimova, E, Kagan, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the uncharacterized protein ABO_0056 from the hydrocarbon-degrading marine bacterium Alcanivorax borkumensis SK2.
TO BE PUBLISHED
|
|
3H1Q
 
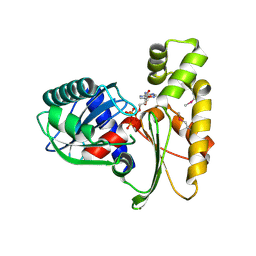 | | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ethanolamine utilization protein EutJ | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Kinney, J, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans
To be Published
|
|
3GQS
 
 | | Crystal structure of the FHA domain of CT664 protein from Chlamydia trachomatis | | Descriptor: | Adenylate cyclase-like protein, PHOSPHATE ION | | Authors: | Majorek, K.A, Cymborowski, M, Chruszcz, M, Evdokimova, E, Egorova, O, Di Leo, R, Zimmerman, M.D, Savchenko, A, Joachimiak, A, Edwards, A.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the FHA domain of CT664 protein from Chlamydia trachomatis
To be Published
|
|
3H1N
 
 | | Crystal structure of probable glutathione S-transferase from Bordetella bronchiseptica RB50 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Probable glutathione S-transferase | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of probable glutathione S-transferase from Bordetella bronchiseptica RB50
To be Published
|
|
3H0K
 
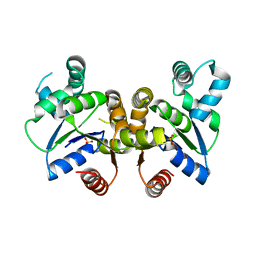 | |
1ILV
 
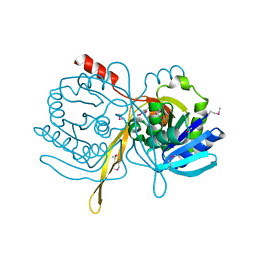 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
4OQJ
 
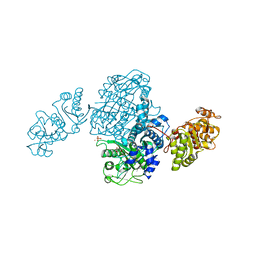 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmQ KS1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PKS, ... | | Authors: | Nocek, B, Mack, J, Endras, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3H04
 
 | | The crystal structure of the protein with unknown function from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | uncharacterized protein | | Authors: | Zhang, R, Tesar, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-08 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the protein with unknown function from Staphylococcus aureus subsp. aureus Mu50
To be Published
|
|
3GL1
 
 | | Crystal structure of ATPase domain of Ssb1 chaperone, a member of the HSP70 family, from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, GLYCEROL, Heat shock protein SSB1, ... | | Authors: | Osipiuk, J, Li, H, Bargassa, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of ATPase domain of Ssb1 chaperone, member of the HSP70 family from Saccharomyces cerevisiae.
To be Published
|
|
4OPE
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS7 | | Descriptor: | NITRATE ION, NRPS/PKS | | Authors: | Osipiuk, J, Mack, J, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3HCY
 
 | | The crystal structure of the domain of putative two-component sensor histidine kinase protein from Sinorhizobium meliloti 1021 | | Descriptor: | Putative two-component sensor histidine kinase protein | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the domain of putative two-component sensor histidine kinase protein from Sinorhizobium meliloti 1021
To be Published
|
|
2M2J
 
 | | Solution NMR structure of the N-terminal domain of STM1478 from Salmonella typhimurium LT2: Target STR147A of the Northeast Structural Genomics consortium (NESG), and APC101565 of the Midwest Center for Structural Genomics (MCSG). | | Descriptor: | Putative periplasmic protein | | Authors: | Houliston, S, Yee, A, Lemak, A, Garcia, M, Wu, B, Savchenko, A, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-21 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
3H3M
 
 | | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica | | Descriptor: | Flagellar protein FliT, UNKNOWN | | Authors: | Shumilin, I.A, Wang, S, Chruszcz, M, Xu, X, Le, B, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-16 | | Release date: | 2009-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica
To be Published
|
|
3H92
 
 | |
3HA9
 
 | | The 1.7A Crystal Structure of a Thioredoxin-like Protein from Aeropyrum pernix | | Descriptor: | uncharacterized Thioredoxin-like protein | | Authors: | Stein, A.J, Cuff, M.E, Sather, A, Hendricks, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-01 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7A Crystal Structure of a Thioredoxin-like Protein from Aeropyrum pernix
To be Published
|
|
4EYO
 
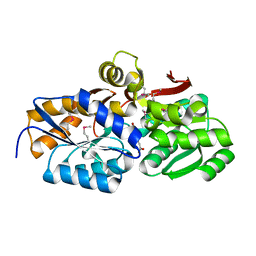 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4Q7O
 
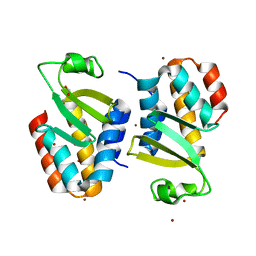 | | The crystal structure of an immunity protein NMB0503 from Neisseria meningitidis MC58 | | Descriptor: | BROMIDE ION, FORMIC ACID, Immunity protein | | Authors: | Tan, K, Stols, L, Eschenfeldt, W, Babnigg, G, Low, D.A, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a contact-dependent growth-inhibition (CDI) immunity protein from Neisseria meningitidis MC58.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
3HH1
 
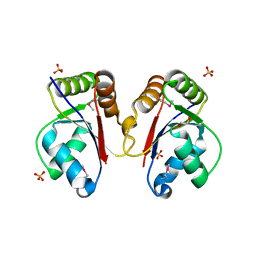 | | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tetrapyrrole methylase family protein | | Authors: | Cuff, M.E, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS.
TO BE PUBLISHED
|
|
4Q6W
 
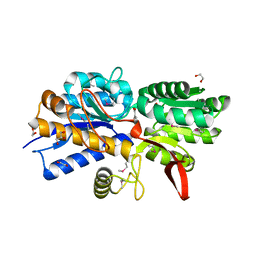 | | Crystal Structure of Periplasmic Binding Protein type 1 from Bordetella pertussis Tohama I complexed with 3-Hydroxy Benzoic Acid | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-23 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Periplasmic Binding Protein type 1 from Bordetella pertussis Tohama I complexed with 3-Hydroxy Benzoic Acid
To be Published, 2014
|
|
3HFI
 
 | | The crystal structure of the putative regulator from Escherichia coli CFT073 | | Descriptor: | Putative regulator | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the putative regulator from Escherichia coli CFT073
To be Published
|
|
3IC5
 
 | |
3IC9
 
 | | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, dihydrolipoamide dehydrogenase | | Authors: | Tan, K, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of dihydrolipoamide dehydrogenase from Colwellia psychrerythraea 34H.
To be Published
|
|
3IR9
 
 | |
