1U5S
 
 | | NMR structure of the complex between Nck-2 SH3 domain and PINCH-1 LIM4 domain | | Descriptor: | Cytoplasmic protein NCK2, PINCH protein, ZINC ION | | Authors: | Vaynberg, J, Fukuda, T, Vinogradova, O, Velyvis, A, Ng, L, Wu, C, Qin, J. | | Deposit date: | 2004-07-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility.
Mol.Cell, 17, 2005
|
|
2I81
 
 | | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced | | Descriptor: | 2-Cys Peroxiredoxin | | Authors: | Artz, J.D, Qiu, W, Dong, A, Lew, J, Ren, H, Zhao, Y, Kozieradski, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Plasmodium vivax 2-Cys Peroxiredoxin, Reduced
To be published
|
|
1U5T
 
 | | Structure of the ESCRT-II endosomal trafficking complex | | Descriptor: | Defective in vacuolar protein sorting; Vps36p, Hypothetical 23.6 kDa protein in YUH1-URA8 intergenic region, appears to be functionally related to SNF7; Snf8p | | Authors: | Hierro, A, Sun, J, Rusnak, A.S, Kim, J, Prag, G, Emr, S.D, Hurley, J.H. | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of ESCRT-II endosomal trafficking complex
Nature, 431, 2004
|
|
2VHK
 
 | |
2V5O
 
 | | STRUCTURE OF HUMAN IGF2R DOMAINS 11-14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, CHLORIDE ION | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
4CXM
 
 | | Crystal Structure of Plasmodium Falciparum Spermidine Synthase in Complex with METHYLTHIOADENOSIN AND SPERMIDINE after catalysis in crystal | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, SPERMIDINE, SPERMIDINE SYNTHASE | | Authors: | Sprenger, J, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2014-04-07 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2IBO
 
 | | X-ray Crystal Structure of Protein SP2199 from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR31 | | Descriptor: | Hypothetical protein SP2199 | | Authors: | Seetharaman, J, Abashidze, M, Forouhar, F, Shastry, R, Conover, K, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-11 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein SP2199 from Streptococcus pneumoniae, Northeast structural genomics target SpR31
To be Published
|
|
8AQ8
 
 | | FAD-dependent monooxygenase from Stenotrophomonas maltophilia | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maly, M, Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tetracycline-modifying enzyme SmTetX from Stenotrophomonas maltophilia.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
1U7C
 
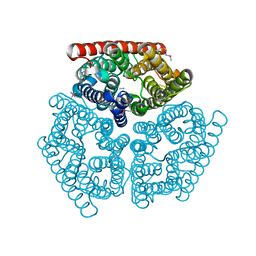 | | Crystal Structure of AmtB from E.Coli with Methyl Ammonium. | | Descriptor: | METHYLAMINE, Probable ammonium transporter | | Authors: | Khademi, S, O'Connell III, J, Remis, J, Robles-Colmenares, Y, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A
Science, 305, 2004
|
|
2I2L
 
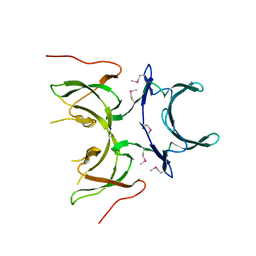 | | X-ray Crystal Structure of Protein yopX from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR411. | | Descriptor: | YopX protein | | Authors: | Vorobiev, S.M, Zhou, W, Seetharaman, J, Forouhar, F, Kuzin, A.A, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-16 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the hypothetical protein yopX from Bacillus subtilis
To be Published
|
|
1UAT
 
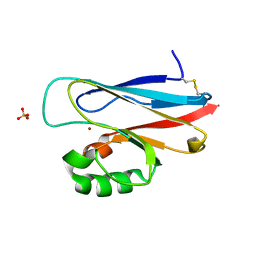 | | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J | | Descriptor: | Azurin iso-2, COPPER (II) ION, SULFATE ION | | Authors: | Inoue, T, Suzuki, S, Nisho, N, Yamaguchi, K, Kataoka, K, Tobari, J, Yong, X, Hamanaka, S, Matsumura, H, Kai, Y. | | Deposit date: | 2003-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J
J.Mol.Biol., 333, 2003
|
|
1KDN
 
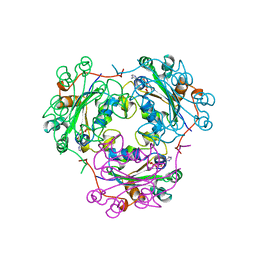 | | STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Cherfils, J, Xu, Y.W, Morera, S, Janin, J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1KLM
 
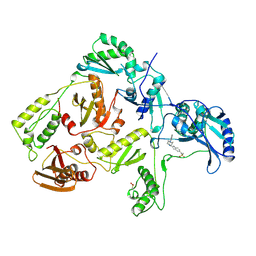 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH BHAP U-90152 | | Descriptor: | (1-(5-METHANSULPHONAMIDO-1H-INDOL-2-YL-CARBONYL)4-[METHYLAMINO)PYRIDINYL]PIPERAZINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R.M, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1997-03-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4CWA
 
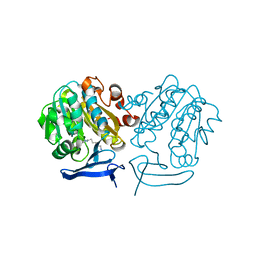 | | Structure of Plasmodium Falciparum Spermidine Synthase in Complex with 1H-Benzimidazole-2-pentanamine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5-(1H-benzimidazol-2-yl)pentan-1-amine, SPERMIDINE SYNTHASE | | Authors: | Sprenger, J, Halander, J.C, Svensson, B, Al-Karadaghi, S, Persson, L. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Three-Dimensional Structures of Plasmodium Falciparum Spermidine Synthase with Bound Inhibitors Suggest New Strategies for Drug Design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6F6R
 
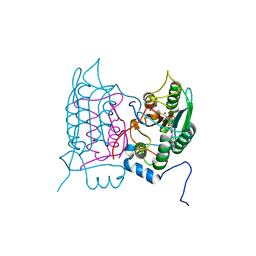 | | Crystal structure of human Caspase-1 with N-{3-[1-((S)-2-Hydroxy-5-oxo-tetrahydro-furan-3-ylcarbamoyl)-ethyl]-1-methyl-2,4-dioxo-1,2,3,4-tetrahydro-pyrimidin-5-yl}-4-(quinoxalin-2-ylamino)-benzamide | | Descriptor: | (3~{S})-3-[[(2~{R})-2-[3-methyl-2,6-bis(oxidanylidene)-5-[[4-(quinoxalin-2-ylamino)phenyl]carbonylamino]pyrimidin-1-yl]propanoyl]amino]-4-oxidanyl-butanoic acid, Caspase-1, SULFATE ION | | Authors: | Fournier, J.F, Clary, L, Chambon, S, Dumais, L, Harris, C.S, Millois-Barbuis, C, Pierre, R, Talano, S, Thoreau, E, Aubert, J, Aurelly, M, Bouix-Peter, C, Brethon, A, Chantalat, L, Christin, O, Comino, C, El-Bazbouz, G, Ghilini, A.L, Isabet, T, Lardy, C, Luzy, A.P, Mathieu, C, Mebrouk, K, Orfila, D, Pascau, J, Reverse, K, Roche, D, Rodeschini, V, Hennequin, L.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Drug Design of Topically Administered Caspase 1 Inhibitors for the Treatment of Inflammatory Acne.
J. Med. Chem., 61, 2018
|
|
2VCJ
 
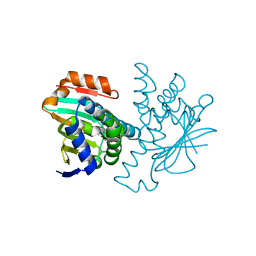 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-(5-chloro-2,4-dihydroxyphenyl)-N-ethyl-4-[4-(morpholin-4-ylmethyl)phenyl]isoxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
1UEF
 
 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
2VCI
 
 | | 4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents for the Treatment of Cancer | | Descriptor: | 5-[2,4-DIHYDROXY-5-(1-METHYLETHYL)PHENYL]-N-ETHYL-4-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]ISOXAZOLE-3-CARBOXAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Aherne, W, Barril, X, Borgognoni, J, Boxal, K, Cansfield, J.E, Cheung, K.M, Collins, I, Davies, N.G.M, Drysdale, M.J, Dymock, B, Eccles, S.A, Finch, H, Fink, A, Hayes, A, Howes, R, Hubbard, R.E, James, K, Jordan, A.M, Lockie, A, Martins, V, Massey, A, Matthews, T.P, McDonald, E, Northfield, C.J, Pearl, L.H, Prodromou, C, Ray, S, Raynaud, F.I, Roughley, S.D, Sharp, S.Y, Surgenor, A, Walmsley, D.L, Webb, P, Wood, M, Workman, P, Wright, L. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4,5-diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer.
J. Med. Chem., 51, 2008
|
|
2IPX
 
 | | Human Fibrillarin | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CALCIUM ION, rRNA 2'-O-methyltransferase fibrillarin | | Authors: | Min, J, Wu, H, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Human Fibrillarin in complex with SAH
To be Published
|
|
1U6V
 
 | | NMR structure of a V3 (IIIB isolate) peptide bound to 447-52D, a human HIV-1 neutralizing antibody | | Descriptor: | V3 peptide | | Authors: | Rosen, O, Chill, J, Sharon, M, Kessler, N, Mester, B, Zolla-Pazner, S, Anglister, J. | | Deposit date: | 2004-08-02 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Induced fit in HIV-neutralizing antibody complexes: evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization.
Biochemistry, 44, 2005
|
|
1U77
 
 | | Crystal Structure of Ammonia Channel AmtB from E. Coli | | Descriptor: | Probable ammonium transporter | | Authors: | Khademi, S, O'Connell III, J, Remis, J, Robles-Colmenares, Y, Miercke, L.J.W, Stroud, R.M. | | Deposit date: | 2004-08-02 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A
Science, 305, 2004
|
|
2VL1
 
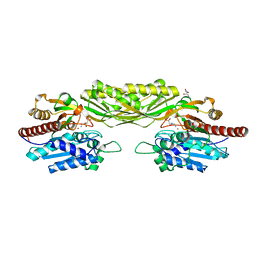 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri in complex with a gly-gly peptide | | Descriptor: | BETA-ALANINE SYNTHASE, GLYCINE, ZINC ION | | Authors: | Andersen, B, Lundgren, S, Dobritzsch, D, Piskur, J. | | Deposit date: | 2008-01-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Recruited Protease is Involved in Catabolism of Pyrimidines.
J.Mol.Biol., 379, 2008
|
|
2J5U
 
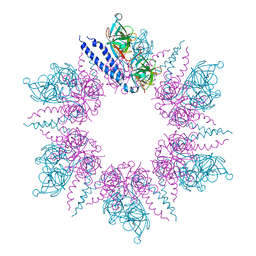 | | MreC Lysteria monocytogenes | | Descriptor: | MREC PROTEIN | | Authors: | van den Ent, F, Leaver, M, Bendezu, F, Errington, J, de Boer, P, Lowe, J. | | Deposit date: | 2006-09-19 | | Release date: | 2006-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric Structure of the Cell Shape Protein Mrec and its Functional Implications.
Mol.Microbiol., 62, 2006
|
|
2VML
 
 | |
