5UMQ
 
 | | Crystal structure of TnmS1, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMX
 
 | | Crystal structure of TnmS3 in complex with riboflavin | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UNC
 
 | | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus | | Descriptor: | FORMIC ACID, L(+)-TARTARIC ACID, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus
To Be Published
|
|
4GYT
 
 | | Crystal structure of lpg0076 protein from Legionella pneumophila | | Descriptor: | SODIUM ION, uncharacterized protein | | Authors: | Michalska, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Crystal structure of lpg0076 protein from Legionella pneumophila
To be Published
|
|
4ERU
 
 | | Crystal Structure of Putative Cytoplasmic Protein, YciF Bacterial Stress Response Protein from Salmonella enterica | | Descriptor: | D-MALATE, MAGNESIUM ION, YciF Bacterial Stress Response Protein | | Authors: | Kim, Y, Wu, R, Jedrzejczak, R, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-20 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Putative Cytoplasmic Protein, YciF Bacterial Stress Response Protein from Salmonella enterica
To be Published
|
|
4EVQ
 
 | | Crystal structure of ABC transporter from R. palustris - solute binding protein (RPA0668) in complex with 4-hydroxybenzoate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Michalska, K, Mack, J.C, Zerbs, S, Collart, F.R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
5UMP
 
 | | Crystal structure of TnmS3, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMW
 
 | | Crystal structure of TnmS2, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
4JGR
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, R131A, from Bacillus subtilis subsp at 2.4A resolution | | Descriptor: | ACETIC ACID, GLYCEROL, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JWO
 
 | | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Tan, K, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-27 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776
To be Published
|
|
5VPJ
 
 | | The crystal structure of a thioesteras from Actinomadura verrucosospora. | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, Thioesterase | | Authors: | Tan, K, Joachimiak, G, Endres, M, Phillips Jr, G.N, Joachmiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-05-05 | | Release date: | 2017-07-19 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of a thioesteras from Actinomadura verrucosospora.
To Be Published
|
|
4HAM
 
 | | Crystal Structure of Transcriptional Antiterminator from Listeria monocytogenes EGD-e | | Descriptor: | GLYCEROL, Lmo2241 protein, SULFATE ION | | Authors: | Kim, Y, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal Structure of Transcriptional Antiterminator from Listeria monocytogenes EGD-e
To be Published
|
|
4FCG
 
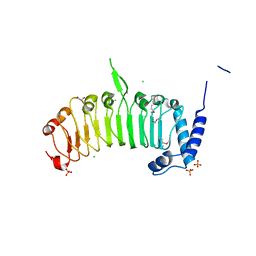 | | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Zimmerman, M.D, Minor, W, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL)
To be Published
|
|
4K08
 
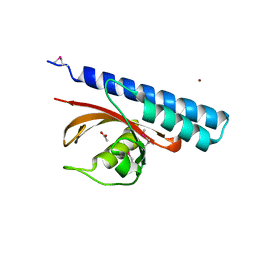 | | Periplasmic sensor domain of chemotaxis protein, Adeh_3718 | | Descriptor: | ACETATE ION, Methyl-accepting chemotaxis sensory transducer, ZINC ION | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-07-17 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
5T87
 
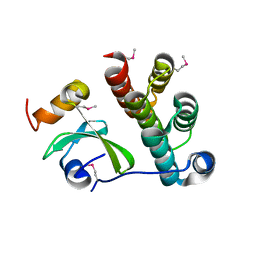 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
4GYU
 
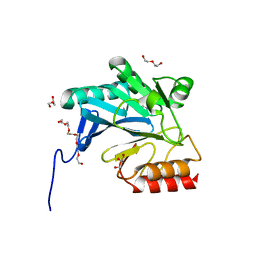 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
4HCJ
 
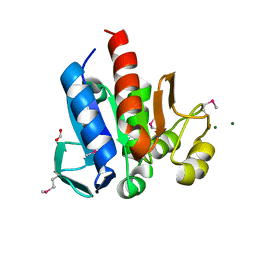 | | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii | | Descriptor: | CHLORIDE ION, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Kim, Y, Bigelow, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii
To be Published
|
|
4EYO
 
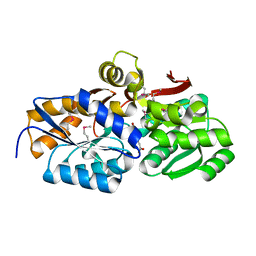 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4I76
 
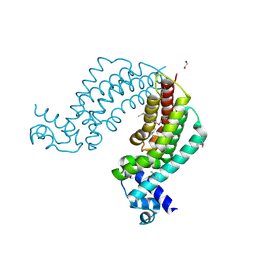 | | Crystal structure of transcriptional regulator TM1030 with octanol | | Descriptor: | 1,2-ETHANEDIOL, OCTAN-1-OL, Transcriptional regulator, ... | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of transcriptional regulator TM1030 with octanol
To be Published
|
|
5VVH
 
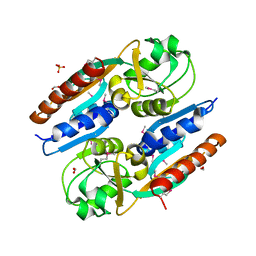 | | Crystal Structure of the Effector Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens | | Descriptor: | FORMIC ACID, Octopine catabolism/uptake operon regulatory protein OccR, SULFATE ION | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-21 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
4HYL
 
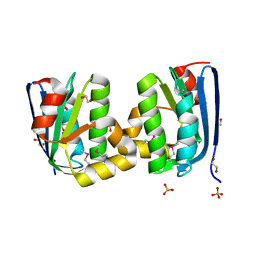 | | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stage II sporulation protein | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-13 | | Release date: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365
To be Published
|
|
4FB4
 
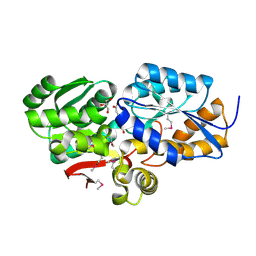 | | The Structure of an ABC-Transporter Family Protein from Rhodopseudomonas palustris in Complex with Caffeic Acid | | Descriptor: | CAFFEIC ACID, GLYCEROL, Putative branched-chain amino acid transport system substrate-binding protein | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-22 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4I66
 
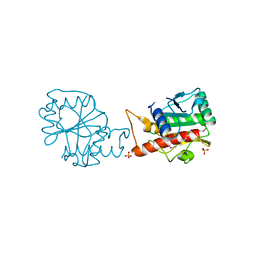 | |
4FC9
 
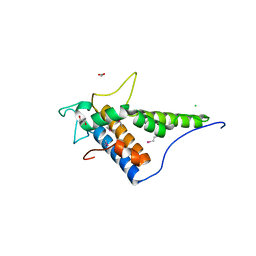 | | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, uncharacterized protein | | Authors: | Singer, A.U, Xu, X, Cui, H, Tan, K, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL)
To be Published
|
|
4E2G
 
 | | Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, Cupin 2 conserved barrel domain protein, NICKEL (II) ION, ... | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-03-08 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of Cupin fold protein Sthe2323 from Sphaerobacter thermophilus
To be Published
|
|
