5KWO
 
 | |
5KWZ
 
 | |
5KX2
 
 | |
5KX0
 
 | |
5KWP
 
 | |
5KX1
 
 | |
5KPH
 
 | |
5KVN
 
 | |
3Q6O
 
 | | Oxidoreductase Fragment of Human QSOX1 | | Descriptor: | SULFATE ION, Sulfhydryl oxidase 1 | | Authors: | Fass, D, Alon, A. | | Deposit date: | 2011-01-03 | | Release date: | 2012-05-30 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The dynamic disulphide relay of quiescin sulphydryl oxidase.
Nature, 488, 2012
|
|
3QD9
 
 | |
3QCP
 
 | | QSOX from Trypanosoma brucei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QSOX from Trypanosoma brucei (TbQSOX) | | Authors: | Alon, A, Fass, D. | | Deposit date: | 2011-01-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The dynamic disulphide relay of quiescin sulphydryl oxidase.
Nature, 488, 2012
|
|
7OIU
 
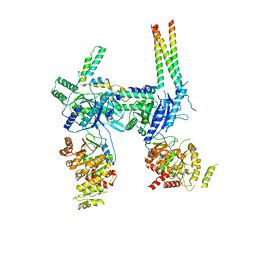 | | Inner Membrane Complex (IMC) protomer structure (TrwM/VirB3, TrwK/VirB4, TrwG/VirB8tails) from the fully-assembled R388 type IV secretion system determined by cryo-EM. | | Descriptor: | TrwG protein, TrwK protein, TrwM protein | | Authors: | Mace, K, Vadakkepat, A.K, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-05-12 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a type IV secretion system.
Nature, 607, 2022
|
|
7Q1V
 
 | |
8AH9
 
 | |
3LEF
 
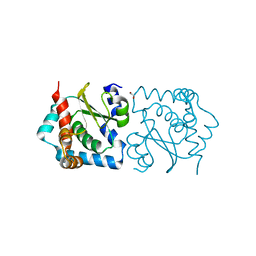 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1Z6NA_001 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein 4E10_S0_1Z6NA_001 (T18) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-14 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
8UOS
 
 | |
8UPB
 
 | |
8UPA
 
 | | Structure of gp130 in complex with a de novo designed IL-6 mimetic | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, De novo designed IL-6 mimetic, Interleukin-6 receptor subunit beta, ... | | Authors: | Borowska, M.T, Jude, K.M, Garcia, K.C. | | Deposit date: | 2023-10-22 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo design of miniprotein antagonists of cytokine storm inducers.
Nat Commun, 15, 2024
|
|
3LF6
 
 | | Crystal structure of HIV epitope-scaffold 4E10_1XIZA_S0_001_N | | Descriptor: | PHOSPHATE ION, Putative phosphotransferase system | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LG7
 
 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1EZ3A_002_C | | Descriptor: | 4E10_S0_1EZ3A_002_C (T246), SULFATE ION | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-19 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LHP
 
 | |
3LH2
 
 | |
3LF9
 
 | | Crystal structure of HIV epitope-scaffold 4E10_D0_1IS1A_001_C | | Descriptor: | 4E10_D0_1IS1A_001_C (T161) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-16 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3MIP
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-8GCG) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*CP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-11 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
3MIS
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-8G) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*CP*TP*CP*CP*G)-3'), ... | | Authors: | Taylor, G.K, Stoddard, B.L. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
