8DG1
 
 | |
8D11
 
 | |
8CXE
 
 | |
8D10
 
 | |
8DG2
 
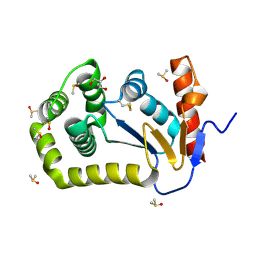 | | Crystal Structure of EcDsbA in a complex with DMSO | | Descriptor: | COPPER (II) ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Whitehouse, R.L, Ilyichova, O.V, Taylor, A.J. | | Deposit date: | 2022-06-23 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment screening libraries for the identification of protein hot spots and their minimal binding pharmacophores.
Rsc Med Chem, 14, 2023
|
|
1HKA
 
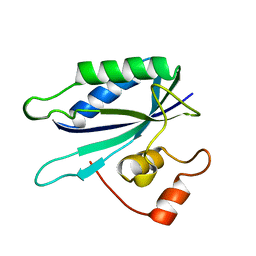 | | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Authors: | Xiao, B, Shi, G, Chen, X, Yan, H, Ji, X. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents.
Structure Fold.Des., 7, 1999
|
|
1LDM
 
 | |
1LVN
 
 | |
1AV4
 
 | |
1AVL
 
 | |
1AVK
 
 | | CRYSTAL STRUCTURES OF THE COPPER-CONTAINING AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS IN THE HOLO-AND APO-FORMS: IMPLICATIONS FOR THE BIOGENESIS OF TOPA QUINONE | | Descriptor: | AMINE OXIDASE | | Authors: | Wilce, M.C.J, Guss, J.M, Freeman, H.C, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 1997-09-16 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone.
Biochemistry, 36, 1997
|
|
1C8Z
 
 | |
8G1U
 
 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
8GEM
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-ethyl-N-({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)-2-(1H-pyrazol-1-yl)ethanamine | | Descriptor: | N-ethyl-N-({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)-2-(1H-pyrazol-1-yl)ethan-1-amine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GDM
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with {[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}(methyl)[1-(thiophen-2-yl)ethyl]amine | | Descriptor: | (1S)-N-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-N-methyl-1-(thiophen-2-yl)ethan-1-amine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-06 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GEY
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with 4-(hydroxymethyl)-1-[(4-methoxy-5,6,7,8-tetrahydronaphthalen-1-yl)sulfonyl]piperidin-4-ol | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(hydroxymethyl)-1-(4-methoxy-5,6,7,8-tetrahydronaphthalene-1-sulfonyl)piperidin-4-ol, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GD2
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-(2-thienylmethyl)methanamine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(thiophen-2-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GEV
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with 1-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-4-(methoxymethyl)piperidine | | Descriptor: | 1-{[3-(diphenylmethyl)-1,2,4-oxadiazol-5-yl]methyl}-4-(methoxymethyl)piperidine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8GEU
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with methyl({3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}methyl)[(1-methylpyrazol-4-yl)methyl]amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-1-{3-[1-(4-methylphenyl)cyclopentyl]-1,2,4-oxadiazol-5-yl}-N-[(1-methyl-1H-pyrazol-4-yl)methyl]methanamine, Retinol-binding protein 1 | | Authors: | Plau, J, Golczak, M. | | Deposit date: | 2023-03-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Nonretinoid Inhibitors of CRBP1: Structural and Dynamic Insights for Ligand-Binding Mechanisms.
Acs Chem.Biol., 18, 2023
|
|
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-12-27 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8HTT
 
 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | Authors: | Han, A, Kim, H.M. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GolpHCat (TMEM87A), a unique voltage-dependent cation channel in Golgi apparatus, contributes to Golgi-pH maintenance and hippocampus-dependent memory.
Nat Commun, 15, 2024
|
|
8JED
 
 | | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors | | Descriptor: | mRNA-capping enzyme regulatory subunit OPG124 | | Authors: | Wang, D, Zhao, R, Shu, W, Hu, W, Wang, M, Cao, J, Zhou, X. | | Deposit date: | 2023-05-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors.
Int.J.Biol.Macromol., 253, 2023
|
|
7JWR
 
 | |
7JX2
 
 | |
7JVG
 
 | | Cellular retinol-binding protein 2 (CRBP2) in complex with 1-arachidonoylglycerol | | Descriptor: | (2S)-2,3-dihydroxypropyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Golczak, M. | | Deposit date: | 2020-08-21 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for the interaction of cellular retinol binding protein 2 (CRBP2) with nonretinoid ligands.
J.Lipid Res., 62, 2021
|
|
