7DDO
 
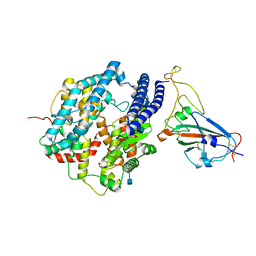 | | Cryo-EM structure of human ACE2 and GD/1/2019 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
7CDW
 
 | |
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
9F99
 
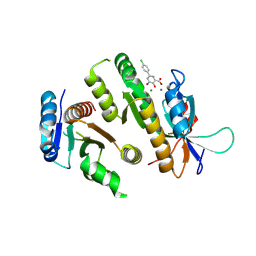 | | Crystal structure of MUS81-EME1 bound by compound 10. | | Descriptor: | 2-(4-chlorophenyl)-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9M
 
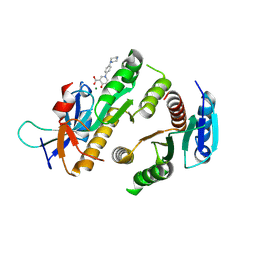 | | Crystal structure of MUS81-EME1 bound by compound 21. | | Descriptor: | 5-oxidanyl-4-oxidanylidene-1-(4-piperazin-1-ylphenyl)pyridine-3-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9A
 
 | | Crystal structure of MUS81-EME1 bound by compound 12. | | Descriptor: | 2-naphthalen-2-yl-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.911 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9K
 
 | | Crystal structure of MUS81-EME1 bound by compound 15. | | Descriptor: | 5-oxidanyl-6-oxidanylidene-2-(4-phenylphenyl)-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F9L
 
 | | Crystal structure of MUS81-EME1 bound by compound 16. | | Descriptor: | 2-[2-[4-(cyanomethyl)phenyl]phenyl]-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9F98
 
 | | Crystal structure of MUS81-EME1, apo form. | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
5XK0
 
 | | Structure of 8-mer DNA2 | | Descriptor: | DNA (5'-D(*GP*CP*CP*CP*GP*AP*GP*C)-3') | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5XM8
 
 | |
5XK1
 
 | | Structure of 8-mer DNA3 | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*CP*CP*CP*C)-3') | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6XRL
 
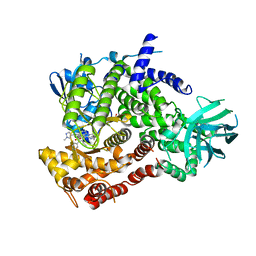 | | Crystal structure of human PI3K-gamma in complex with inhibitor IPI-549 | | Descriptor: | 2-amino-N-[(1S)-1-{8-[(1-methyl-1H-pyrazol-4-yl)ethynyl]-1-oxo-2-phenyl-1,2-dihydroisoquinolin-3-yl}ethyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
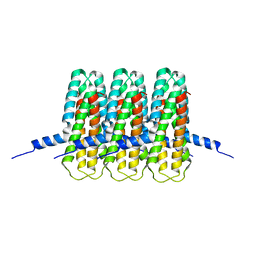
8CQR
 
 | |
4N7M
 
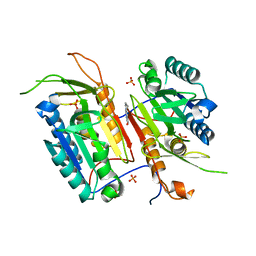 | |
4N6G
 
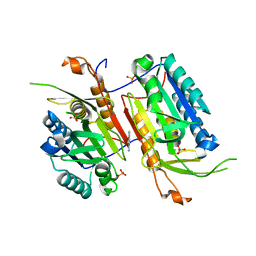 | |
4NBK
 
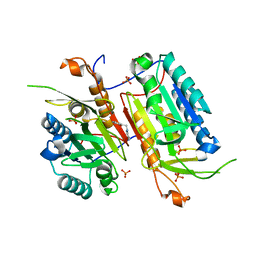 | |
7NHR
 
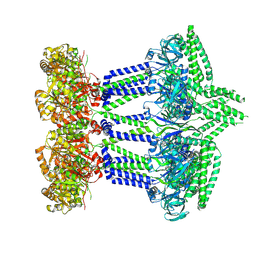 | |
7NIH
 
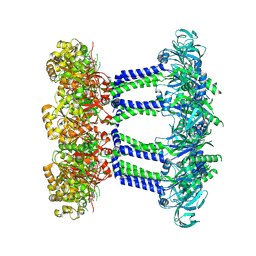 | | Wzc-K540M MgADP C8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NHS
 
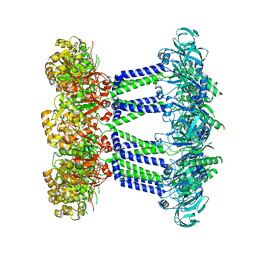 | | Wzc K540M C8 | | Descriptor: | Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
7NII
 
 | | Wzc-K540M MgADP C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Naismith, J.H, Liu, J.W, Yang, Y. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | The molecular basis of regulation of bacterial capsule assembly by Wzc.
Nat Commun, 12, 2021
|
|
