2KZD
 
 | | Structure of a (3+1) G-quadruplex formed by hTERT promoter sequence | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*IP*AP*GP*GP*GP*GP*CP*TP*GP*GP*GP*AP*GP*GP*GP*C)-3') | | Authors: | Lim, K.W, Lacroix, L, Yue, D.J.E, Lim, J.K.C, Lim, J.M.W, Phan, A.T. | | Deposit date: | 2010-06-16 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two distinct G-quadruplex conformations in the hTERT promoter
J.Am.Chem.Soc., 132, 2010
|
|
2GV4
 
 | | Solution structure of the poliovirus 3'-UTR Y-stem | | Descriptor: | Short RNA strand 5'-GGACCUCUCGAAAGAGUGGUCC-3' | | Authors: | Heus, H.A, Zoll, J, Tessari, M, van Kuppeveld, F.J.M, Melchers, W.J.G. | | Deposit date: | 2006-05-02 | | Release date: | 2007-03-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Breaking pseudo-twofold symmetry in the poliovirus 3'-UTR Y-stem by restoring Watson-Crick base pairs.
Rna, 13, 2007
|
|
2KZE
 
 | | Structure of an all-parallel-stranded G-quadruplex formed by hTERT promoter sequence | | Descriptor: | DNA (5'-D(*AP*IP*GP*GP*GP*AP*GP*GP*GP*IP*CP*TP*GP*GP*GP*AP*GP*GP*GP*C)-3') | | Authors: | Lim, K.W, Lacroix, L, Yue, D.J.E, Lim, J.K.C, Lim, J.M.W, Phan, A.T. | | Deposit date: | 2010-06-16 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two distinct G-quadruplex conformations in the hTERT promoter
J.Am.Chem.Soc., 132, 2010
|
|
1GUV
 
 | | Structure of human chitotriosidase | | Descriptor: | 1,2-ETHANEDIOL, CHITOTRIOSIDASE | | Authors: | Von Moeller, H, Houston, D, Boot, R.G, Aerts, J.M.F.G, Van Aalten, D.M.F. | | Deposit date: | 2002-01-31 | | Release date: | 2003-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Human Chitotriosidase - Implications for Specific Inhibitor Design and Function of Mammalian Chitinase-Like Lectins
J.Biol.Chem., 277, 2002
|
|
1HG3
 
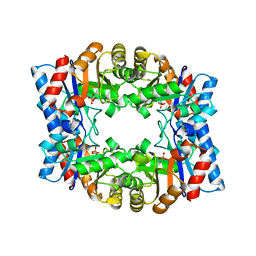 | | Crystal structure of tetrameric TIM from Pyrococcus woesei. | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Walden, H, Bell, G.S, Russell, R.J.M, Siebers, B, Hensel, R, Taylor, G.L. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tiny Tim: A Small, Tetrameric, Hyperthermostable Triosephosphate Isomerase
J.Mol.Biol., 306, 2001
|
|
1HKI
 
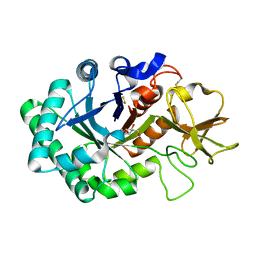 | | Crystal structure of human chitinase in complex with glucoallosamidin B | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOTRIOSIDASE, METHYL N-ACETYL ALLOSAMINE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
1HKJ
 
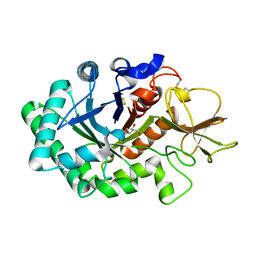 | | Crystal structure of human chitinase in complex with methylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITOTRIOSIDASE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
1HKK
 
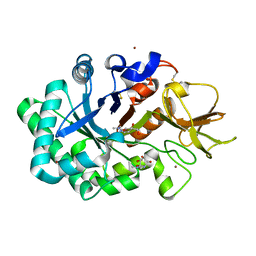 | | High resoultion crystal structure of human chitinase in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITOTRIOSIDASE-1, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase.
J.Biol.Chem., 278, 2003
|
|
1HKM
 
 | | High resolution crystal structure of human chitinase in complex with demethylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, CHITOTRIOSIDASE, METHYL N-ACETYL ALLOSAMINE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
1ARR
 
 | | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF THE ARC REPRESSOR | | Descriptor: | ARC REPRESSOR | | Authors: | Bonvin, A.M.J.J, Vis, H, Burgering, M.J.M, Breg, J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-08-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations.
J.Mol.Biol., 236, 1994
|
|
1ARQ
 
 | | RELAXATION MATRIX REFINEMENT OF THE SOLUTION STRUCTURE OF THE ARC REPRESSOR | | Descriptor: | ARC REPRESSOR | | Authors: | Bonvin, A.M.J.J, Vis, H, Burgering, M.J.M, Breg, J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-08-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations.
J.Mol.Biol., 236, 1994
|
|
8D5J
 
 | |
5NJN
 
 | | Roll out the beta-barrel: structure and mechanism of Pac13, a unique nucleoside dehydratase | | Descriptor: | Putative cupin_2 domain-containing isomerase | | Authors: | Michailidou, F, Bent, A.F, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2017-03-29 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1B2P
 
 | | NATIVE MANNOSE-SPECIFIC BULB LECTIN FROM SCILLA CAMPANULATA (BLUEBELL) AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (LECTIN) | | Authors: | Wood, S.D, Wright, L.M, Reynolds, C.D, Rizkallah, P.J, Allen, A.K, Peumans, W.J, Van Damme, E.J.M. | | Deposit date: | 1998-11-30 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the native (unligated) mannose-specific bulb lectin from Scilla campanulata (bluebell) at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5NJO
 
 | | Roll out the beta-barrel: structure and mechanism of Pac13, a unique nucleoside dehydratase | | Descriptor: | Putative cupin_2 domain-containing isomerase | | Authors: | Michailidou, F, Bent, A.F, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2017-03-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pac13 is a Small, Monomeric Dehydratase that Mediates the Formation of the 3'-Deoxy Nucleoside of Pacidamycins.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6QGM
 
 | | VirX1 apo structure | | Descriptor: | VirX1 | | Authors: | Gkotsi, D.S, Ludewig, H, Sharma, S.V, Unsworth, W.P, Taylor, R.J.K, McLachlan, M.M.W, Shanahan, S, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A marine viral halogenase that iodinates diverse substrates.
Nat.Chem., 11, 2019
|
|
8FHL
 
 | |
8FHU
 
 | |
6Z4A
 
 | |
7K14
 
 | | Ternary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN and methanesulfonate | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-09-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JYB
 
 | | Binary soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P, El Saudi, I.M. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JW9
 
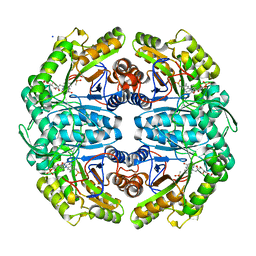 | | Ternary cocrystal structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens | | Descriptor: | Alkanesulfonate monooxygenase, FLAVIN MONONUCLEOTIDE, SODIUM ION, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
7JV3
 
 | |
7K64
 
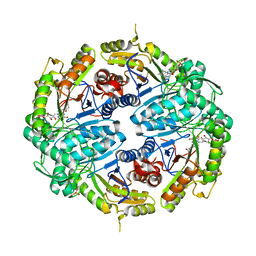 | | Binary titrated soak structure of alkanesulfonate monooxygenase MsuD from Pseudomonas fluorescens with FMN | | Descriptor: | Alkanesulfonate monooxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liew, J.J.M, Dowling, D.P. | | Deposit date: | 2020-09-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the alkanesulfonate monooxygenase MsuD provide insight into C-S bond cleavage, substrate scope, and an unexpected role for the tetramer.
J.Biol.Chem., 297, 2021
|
|
6F4C
 
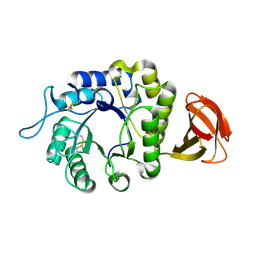 | | Nicotiana benthamiana alpha-galactosidase | | Descriptor: | alpha-galactosidase | | Authors: | Kytidou, K, Aerts, J.M.F.G, Pannu, N.S. | | Deposit date: | 2017-11-29 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nicotiana benthamianaalpha-galactosidase A1.1 can functionally complement human alpha-galactosidase A deficiency associated with Fabry disease.
J. Biol. Chem., 293, 2018
|
|
