3TZD
 
 | | Crystal structure of the complex of Human Chromobox Homolog 3 (CBX3) | | Descriptor: | Chromobox protein homolog 3, Histone H1.4 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
4HK4
 
 | | Crystal structure of apo Tyrosine-tRNA ligase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal structure of apo Tyrosine-tRNA ligase mutant protein
To be Published
|
|
4HPW
 
 | | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine | | Descriptor: | 3-methoxy-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine
To be Published
|
|
2RJD
 
 | | Crystal structure of L3MBTL1 protein | | Descriptor: | Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RJC
 
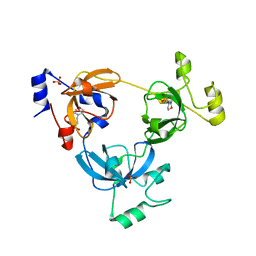 | | Crystal structure of L3MBTL1 protein in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RJF
 
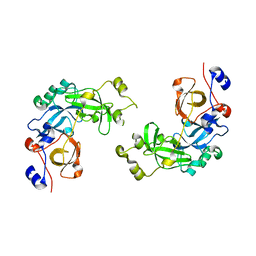 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 12-30), orthorhombic form I | | Descriptor: | Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RJE
 
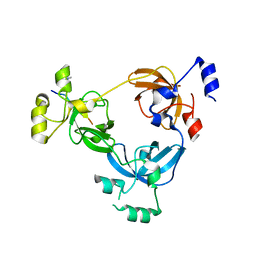 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), orthorhombic form II | | Descriptor: | CHLORIDE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2PQW
 
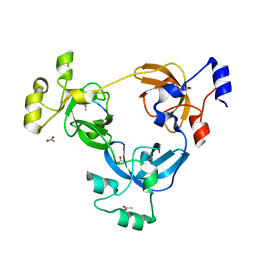 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), trigonal form | | Descriptor: | ACETATE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-02 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2KEO
 
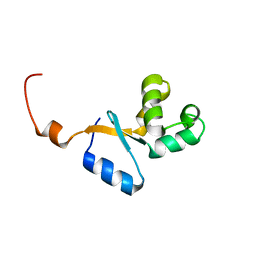 | | Solution NMR structure of human protein HS00059, cytochrome-b5-like domain of the HERC2 E3 ligase. Northeast structural genomics consortium (NESG) target ht98a | | Descriptor: | Probable E3 ubiquitin-protein ligase HERC2 | | Authors: | Lemak, A, Gutmanas, A, Fares, C, Quyang, H, Li, Y, Montelione, G, Arrowsmith, C, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of human protein HS00059
To be Published
|
|
7XYF
 
 | |
7XYG
 
 | |
7XXE
 
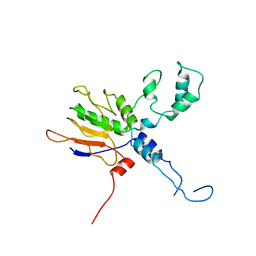 | |
2K28
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 4 | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Fares, C, Gutmanas, A, Ravichandran, M, Loppnau, P, Bountra, C, Weigelt, J, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromo domain of the chromobox protein homolog 4.
To be Published
|
|
2K1B
 
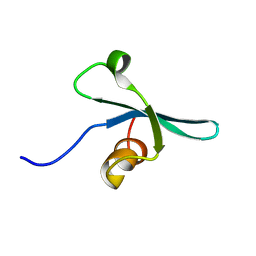 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 7 | | Descriptor: | Chromobox protein homolog 7 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Gutmanas, A, Fares, C, Bountra, C, Weigelt, J, Loppnau, P, Ravichandran, M, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromobox protein homolog 7.
To be Published
|
|
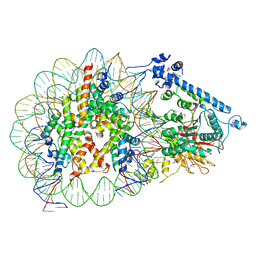
7JY7
 
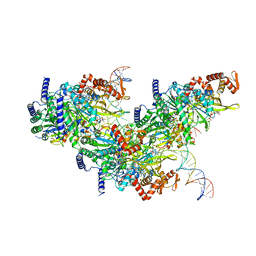 | | Structure of a 12 base pair RecA-D loop complex | | Descriptor: | DNA (27-MER), DNA (48-MER), MAGNESIUM ION, ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2020-08-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of strand exchange from RecA-DNA synaptic and D-loop structures.
Nature, 586, 2020
|
|
7JY9
 
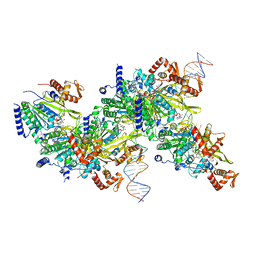 | | Structure of a 9 base pair RecA-D loop complex | | Descriptor: | DNA (27-MER), DNA (42-MER), MAGNESIUM ION, ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2020-08-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of strand exchange from RecA-DNA synaptic and D-loop structures.
Nature, 586, 2020
|
|
7JY6
 
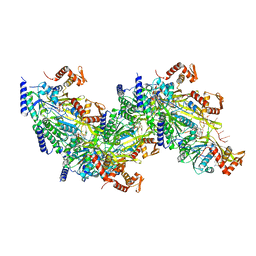 | |
7JY8
 
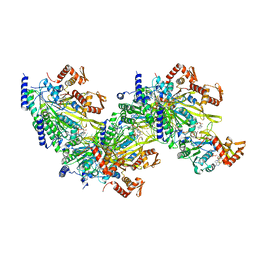 | |
1IYJ
 
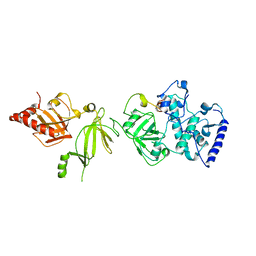 | | STRUCTURE OF A BRCA2-DSS1 COMPLEX | | Descriptor: | Deleted in split hand/split foot protein 1, breast cancer susceptibility | | Authors: | Pavletich, N.P, Jeffrey, P.D, Yang, H.J. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
6P3Y
 
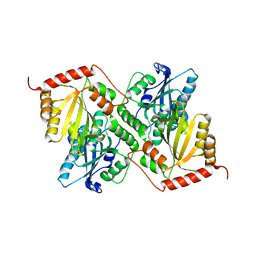 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.4) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6P40
 
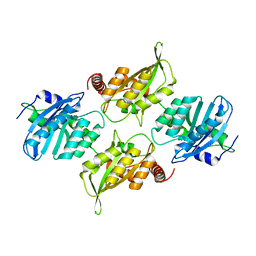 | | Crystal Structure of Full Length APOBEC3G FKL | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
3MF5
 
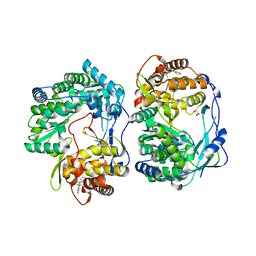 | |
6P3X
 
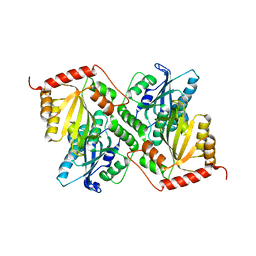 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.0) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
6P3Z
 
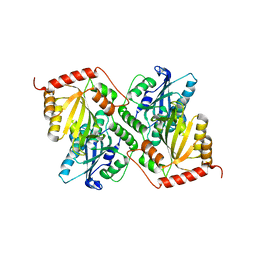 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 5.2) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
8EDJ
 
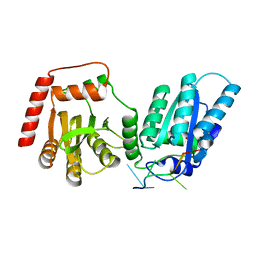 | | Crystal structure of rA3G-ssRNA-GA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*GP*AP*UP*UP*U)-3'), SULFATE ION, ... | | Authors: | Pacheco, J, Yang, H.J, Li, S.-X, Chen, X.S. | | Deposit date: | 2022-09-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
