6UL5
 
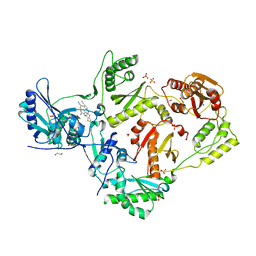 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile (24b), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Pilch, A, Arnold, E. | | Deposit date: | 2019-10-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Characterization of Fluorine-Substituted Diarylpyrimidine Derivatives as Novel HIV-1 NNRTIs with Highly Improved Resistance Profiles and Low Activity for the hERG Ion Channel.
J.Med.Chem., 63, 2020
|
|
3KU0
 
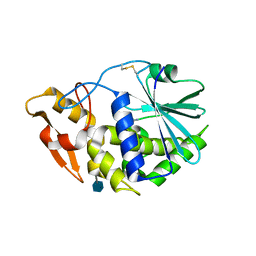 | | Structure of GAP31 with adenine at its binding pocket | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
6ABO
 
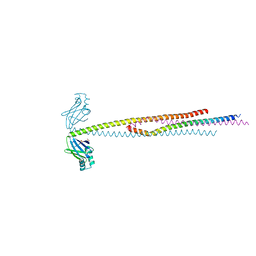 | | human XRCC4 and IFFO1 complex | | Descriptor: | DNA repair protein XRCC4, GLYCEROL, Intermediate filament family orphan 1, ... | | Authors: | Li, J, Liu, L, Liang, H, Liu, Y, Xu, D. | | Deposit date: | 2018-07-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis.
Nat.Cell Biol., 21, 2019
|
|
6VUM
 
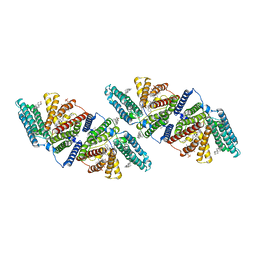 | |
3KTZ
 
 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
8HN6
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | Heavy chain of monoclonal antibody 3G10, Light chain of monoclonal antibody 3G10, Spike protein S1 | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
8HN7
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 3C11, Light chain of monoclonal antibody 3C11, ... | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
1WVJ
 
 | | Exploring the GluR2 ligand-binding core in complex with the bicyclic AMPA analogue (S)-4-AHCP | | Descriptor: | 3-(3-HYDROXY-7,8-DIHYDRO-6H-CYCLOHEPTA[D]ISOXAZOL-4-YL)-L-ALANINE, GLYCEROL, SULFATE ION, ... | | Authors: | Nielsen, B.B, Pickering, D.S, Greenwood, J.R, Brehm, L, Gajhede, M, Schousboe, A, Kastrup, J.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploring the GluR2 ligand-binding core in complex with the bicyclical AMPA analogue (S)-4-AHCP
FEBS J., 272, 2005
|
|
3W2D
 
 | | Crystal Structure of Staphylococcal Eenterotoxin B in complex with a novel neutralization monoclonal antibody Fab fragment | | Descriptor: | Enterotoxin type B, Monoclonal Antibody 3E2 Fab figment heavy chain, Monoclonal Antibody 3E2 Fab figment light chain, ... | | Authors: | Liang, S.Y, Hu, S, Dai, J.X, Guo, Y.J, Lou, Z.Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the neutralization and specificity of Staphylococcal enterotoxin B against its MHC Class II binding site.
MAbs, 6, 2014
|
|
6CDY
 
 | | Crystal structure of TEAD complexed with its inhibitor | | Descriptor: | 2-[(4H-1,2,4-triazol-3-yl)sulfanyl]-N-{4-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]phenyl}acetamide, Transcriptional enhancer factor TEF-4 | | Authors: | LIU, S, HAN, X, LUO, X. | | Deposit date: | 2018-02-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription.
Cell Stem Cell, 26, 2020
|
|
8XDA
 
 | | Cryo-EM structure of urea bound human urea transporter A2. | | Descriptor: | UREA, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J, Zhizheng, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDH
 
 | |
8XDF
 
 | |
8XD9
 
 | |
8XDG
 
 | |
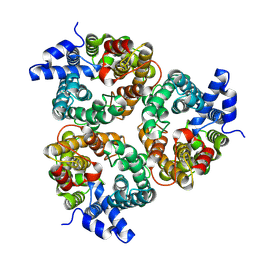
8XDE
 
 | |
8XDD
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | 1-(3-methoxyphenyl)methanamine, 8-hydroxyquinoline-2-carboxylic acid, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8Z0R
 
 | | Cryo-EM structure of tetramer HtmB2-CT | | Descriptor: | special condensation domain in NRPS | | Authors: | Sun, Y.H, Zhang, Z.Y, Mei, Q. | | Deposit date: | 2024-04-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Formation of the Diketopiperazine Moiety by a Distinct Condensation-Like Domain in Hangtaimycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8Z0Q
 
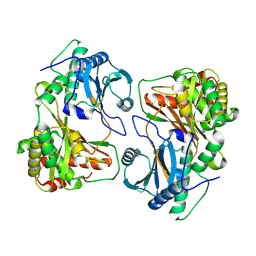 | | Cryo-EM structure of dimer HtmB2-CT | | Descriptor: | Special condensation domain in NRPS | | Authors: | Sun, Y.H, Zhang, Z.Y, Mei, Q. | | Deposit date: | 2024-04-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Formation of the Diketopiperazine Moiety by a Distinct Condensation-Like Domain in Hangtaimycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8Z0S
 
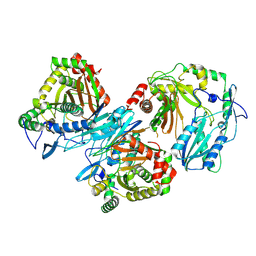 | | Cryo-EM structure of trimer HtmB2-CT | | Descriptor: | special condensation domain in NRPS | | Authors: | Sun, Y.H, Zhang, Z.Y, Mei, Q. | | Deposit date: | 2024-04-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Formation of the Diketopiperazine Moiety by a Distinct Condensation-Like Domain in Hangtaimycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
1FTJ
 
 | |
1FTK
 
 | |
1FTM
 
 | |
1FW0
 
 | |
1FTO
 
 | |
