7HFR
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0005997 | | Descriptor: | (3aS,6S,6aR)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydrocyclopenta[b]pyrrol-6-ol, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
7HFD
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-0004127 | | Descriptor: | 2-[(1S,3aR,7aS)-2-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydro-1H-isoindol-1-yl]propan-2-ol, 2-[(1S,3aS,7aR)-2-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydro-1H-isoindol-1-yl]propan-2-ol, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
7HFT
 
 | |
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7M95
 
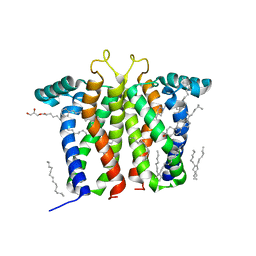 | | Bovine sigma-2 receptor bound to Z1241145220 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[1-(3-phenylpropyl)-1,2,3,6-tetrahydropyridin-4-yl]-1H-pyrrolo[2,3-b]pyridine, CHOLESTEROL, ... | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7MFI
 
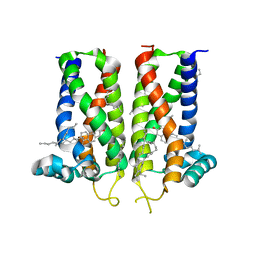 | | Bovine sigma-2 receptor bound to cholesterol | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-04-09 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7M96
 
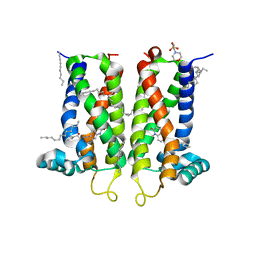 | | Bovine sigma-2 receptor bound to Z4857158944 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-({[(1S)-1-hydroxy-2-methyl-1-phenylpropan-2-yl]amino}methyl)-1-methyl-3,4-dihydroquinolin-2(1H)-one, ... | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7M94
 
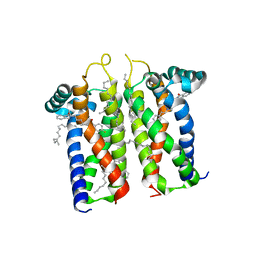 | | Bovine sigma-2 receptor bound to Roluperidone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Roluperidone, Sigma intracellular receptor 2 | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7M93
 
 | | Bovine sigma-2 receptor bound to PB28 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PB28, Sigma intracellular receptor 2 | | Authors: | Alon, A, Kruse, A.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of the sigma 2 receptor enable docking for bioactive ligand discovery.
Nature, 600, 2021
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
9DQH
 
 | | CryoEM structure of Gq-coupled MRGPRD with a new agonist EP-2825 | | Descriptor: | 2-({1-[2-(4-chlorophenyl)-2-methylpropanoyl]piperidin-4-yl}amino)-5,6,7,8-tetrahydroquinazolin-4(3H)-one, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Wang, C, Liu, Y, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-09-24 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-affinity agonists reveal recognition motifs for the MRGPRD GPCR.
Cell Rep, 43, 2024
|
|
9D6H
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-1504 | | Descriptor: | (2R)-3-methyl-2-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]butan-1-ol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
9D6B
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodmain in complex with AVI-607 | | Descriptor: | 2-[(2R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-2-yl]propan-2-ol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-14 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
9D6G
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-3716 | | Descriptor: | DIMETHYL SULFOXIDE, Non-structural protein 3, [(2R,3S)-3-methyl-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-2-yl]methanol | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
9D6I
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with AVI-4317 | | Descriptor: | (3R)-3-hydroxy-3-{[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]methyl}-1lambda~6~-thiane-1,1-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2024-08-15 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Exploration of structure-activity relationships for the SARS-CoV-2 macrodomain from shape-based fragment linking and active learning.
Sci Adv, 11, 2025
|
|
9C1P
 
 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '6218 | | Descriptor: | (5R)-N-[2-(1,2-benzothiazol-3-yl)ethyl]-1-methyl-2,3,4,5-tetrahydro-1H-1-benzazepin-5-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-29 | | Release date: | 2024-10-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
9C2F
 
 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '54149 | | Descriptor: | (1R)-1-(2H-1,3-benzodioxol-4-yl)-N-[2-(1,2-benzothiazol-3-yl)ethyl]ethan-1-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
9DQJ
 
 | | CryoEM structure of Gq-coupled MRGPRD with a new agonist EP-3945 | | Descriptor: | 2-({1-[1-(4-methoxyphenyl)cyclopropane-1-carbonyl]piperidin-4-yl}amino)quinazolin-4(3H)-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Wang, C, Liu, Y, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-09-24 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-affinity agonists reveal recognition motifs for the MRGPRD GPCR.
Cell Rep, 43, 2024
|
|
4M51
 
 | | Crystal structure of amidohydrolase nis_0429 (ser145ala mutant) from nitratiruptor sp. sb155-2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase family protein, BENZOIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Gobble, A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.
Biochemistry, 52, 2013
|
|
4M8T
 
 | |
4NVK
 
 | | Predicting protein conformational response in prospective ligand discovery. | | Descriptor: | Cytochrome c peroxidase, N~2~,N~2~-diethylquinazoline-2,4-diamine, PHOSPHATE ION, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2013-12-05 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery.
Nat Chem, 6, 2014
|
|
4NVE
 
 | |
