1P2O
 
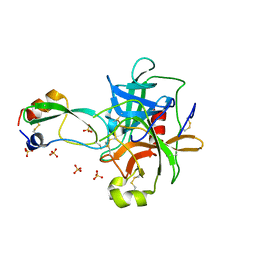 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | Chymotrypsinogen A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2M
 
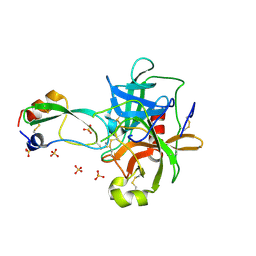 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | Chymotrypsinogen A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2K
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2Q
 
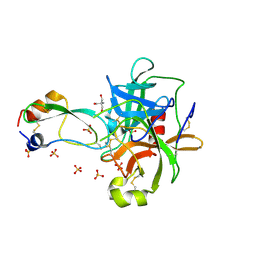 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chymotrypsinogen A, Pancreatic trypsin inhibitor, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
2K37
 
 | | CsmA | | Descriptor: | Chlorosome Protein A | | Authors: | Pedersen, M, Dittmer, J, Underhaug, J, Miller, M, Nielsen, N.C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of CsmA: A small antenna protein from the green sulfur bacterium Chlorobium tepidum.
Febs Lett., 582, 2008
|
|
3DN5
 
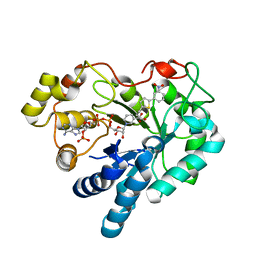 | | Aldose Reductase in complex with novel biarylic inhibitor | | Descriptor: | 3-[5-(3-nitrophenyl)thiophen-2-yl]propanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Klebe, G. | | Deposit date: | 2008-07-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Optimization of Aldose Reductase Inhibitors Originating from Virtual Screening
Chemmedchem, 4, 2009
|
|
2POX
 
 | |
6MD9
 
 | |
6MDA
 
 | |
6MD7
 
 | |
6YO6
 
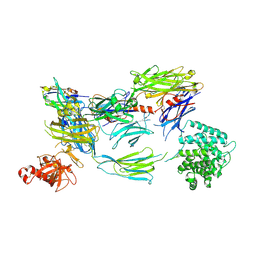 | | Structure of iC3b1 | | Descriptor: | hC3Nb1, iC3b1 alpha chain, iC3b1 beta chain | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2020-04-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Complement Receptor 3 Forms a Compact High-Affinity Complex with iC3b.
J Immunol., 206, 2021
|
|
7P2D
 
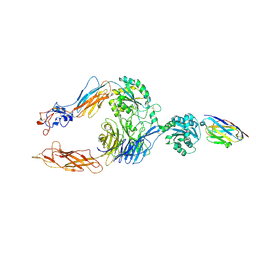 | | Structure of alphaMbeta2/Cd11bCD18 headpiece in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2021-07-05 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the function-modulating effects of nanobody binding to the integrin receptor alpha M beta 2.
J.Biol.Chem., 298, 2022
|
|
6ZFM
 
 | | Structure of alpha-Cobratoxin with a peptide inhibitor | | Descriptor: | 3-[2-[2-[2-[2-[2-(2-azanylethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]propan-1-ol, Alpha-cobratoxin, PENTAETHYLENE GLYCOL, ... | | Authors: | Kiontke, S, Kummel, D. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide Inhibitors of the alpha-Cobratoxin-Nicotinic Acetylcholine Receptor Interaction.
J.Med.Chem., 63, 2020
|
|
3G6W
 
 | |
6TM0
 
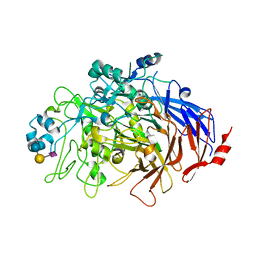 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 6'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
4UX9
 
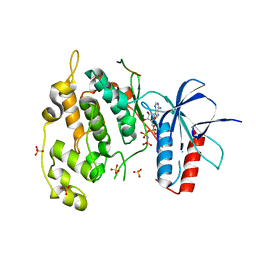 | | Crystal structure of JNK1 bound to a MKK7 docking motif | | Descriptor: | DUAL SPECIFICITY MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7, MITOGEN-ACTIVATED PROTEIN KINASE 8, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kragelj, J, Palencia, A, Nanao, M.H, Maurin, D, Bouvignies, G, Blackledge, M, Ringkjobing-Jensen, M. | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Dynamics of the Mkk7-Jnk Signaling Complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
7ROY
 
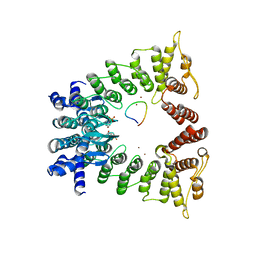 | | The structure of the Fem1B:FNIP1 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Folliculin-interacting protein 1, Protein fem-1 homolog B, ... | | Authors: | Gee, C.L, Mena, E.L, Manford, A.G, Rape, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis and regulation of the reductive stress response.
Cell, 184, 2021
|
|
4EMV
 
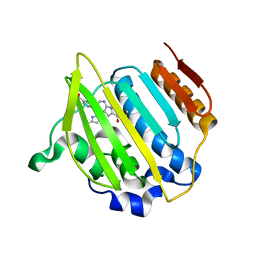 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 5-{2-(ethylcarbamoyl)-4-[3-(trifluoromethyl)-1H-pyrazol-1-yl]-1H-pyrrolo[2,3-b]pyridin-5-yl}pyridine-3-carboxylic acid, DNA topoisomerase IV, B subunit | | Authors: | Boriack-Sjodin, P.A, Manchester, J, Hull, K. | | Deposit date: | 2012-04-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a novel azaindole class of antibacterial agents targeting the ATPase domains of DNA gyrase and Topoisomerase IV.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6DCW
 
 | |
3TH5
 
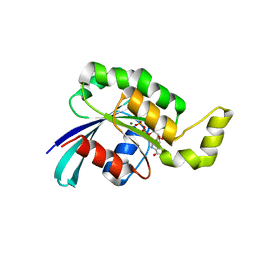 | | Crystal structure of wild-type RAC1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
5W6W
 
 | |
3T3M
 
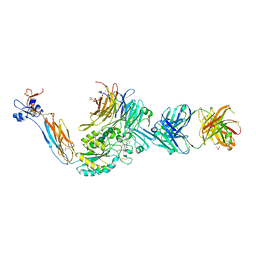 | | A Novel High Affinity Integrin alphaIIbbeta3 Receptor Antagonist That Unexpectedly Displaces Mg2+ from the beta3 MIDAS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2011-07-25 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Design of a High-Affinity Platelet Integrin alphaIIbbeta3 Receptor Antagonist That Disrupts Mg2+ Binding to the MIDAS
Sci Transl Med, 4, 2012
|
|
2E0O
 
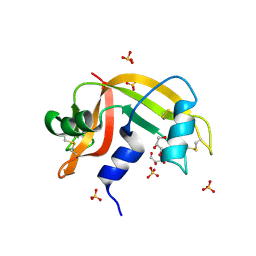 | | Mutant Human Ribonuclease 1 (V52L, D53L, N56L, F59L) | | Descriptor: | GLYCEROL, Ribonuclease, SULFATE ION | | Authors: | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
2E0L
 
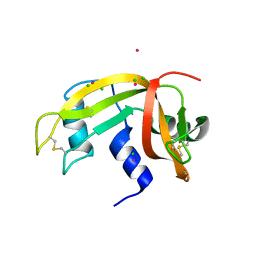 | | Mutant Human Ribonuclease 1 (Q28L, R31L, R32L) | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ribonuclease | | Authors: | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
