4PUD
 
 | | Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylopentaose in active site | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, ZINC ION, ... | | Authors: | Dann, R.D, Solomon, H.V, Lansky, S, Ben-David, A, Lavid, N, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2014-03-13 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Extracellulr Xylanase from Geobacillus stearothermophilus: E159Q mutant, with xylopentaose in active site.
To be Published
|
|
4PRW
 
 | | Xylanase T6 (XT6) from Geobacillus Stearothermophilus in complex with xylohexaose | | Descriptor: | CHLORIDE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Dann, R.D, Solomon, H.V, Lansky, S, Ben-David, A, Lavid, N, Salama, R, Shoham, Y, Shoham, G. | | Deposit date: | 2014-03-06 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Xylanase T6 (XT6) from Geobacillus Stearothermophilus in complex with xylohexaose
To be Published
|
|
2XXI
 
 | | Crystal structure of 1-((4-(3-(trifluoromethyl)-6,7-dihydropyrano(4,3- c(pyrazol-1(4H)-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.6A resolution. | | Descriptor: | 1-({4-[3-(TRIFLUOROMETHYL)-6,7-DIHYDROPYRANO[4,3-C]PYRAZOL-1(4H)-YL]PHENYL}METHYL)-2-PYRROLIDINONE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX9
 
 | | Crystal structure of 1-((2-fluoro-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
4GS5
 
 | | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein, IODIDE ION | | Authors: | Tan, K, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-12 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053
To be Published
|
|
4PIB
 
 | | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uncharacterized Conserved Protein PixA from Burkholderia thailandensis
To Be Published
|
|
4H7N
 
 | |
4H3T
 
 | | Crystal structure of CRISPR-associated protein Cse1 from Acidimicrobium ferrooxidans | | Descriptor: | CRISPR-associated protein, Cse1 family, GLYCEROL | | Authors: | Michalska, K, Stols, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of CRISPR-associated protein Cse1 from Acidimicrobium ferrooxidans
To be Published
|
|
4MPS
 
 | | Crystal structure of rat Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GAL1), Northeast Structural Genomics Consortium Target RnR367A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactoside alpha-2,6-sialyltransferase 1 | | Authors: | Forouhar, F, Meng, L, Milaninia, S, Seetharaman, J, Su, M, Kornhaber, G, Montelione, G.T, Hunt, J.F, Moremen, K.W, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-13 | | Release date: | 2013-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic Basis for N-Glycan Sialylation: STRUCTURE OF RAT alpha 2,6-SIALYLTRANSFERASE (ST6GAL1) REVEALS CONSERVED AND UNIQUE FEATURES FOR GLYCAN SIALYLATION.
J.Biol.Chem., 288, 2013
|
|
4NAS
 
 | | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446.
To be Published
|
|
4HG2
 
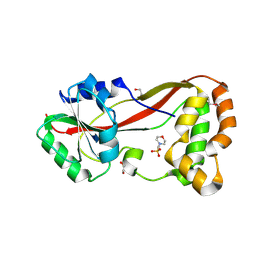 | | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Methyltransferase type 11 | | Authors: | Cuff, M.E, Chhor, G, Clancy, S, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans.
TO BE PUBLISHED
|
|
2XX7
 
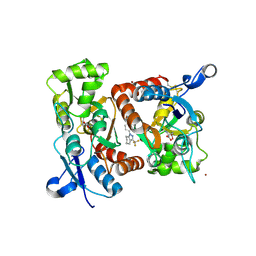 | | Crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | 1-[4-(1-PYRROLIDINYLCARBONYL)PHENYL]-3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XXH
 
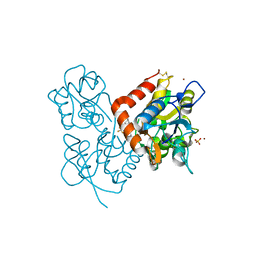 | | Crystal structure of 1-(4-(2-oxo-2-(1-pyrrolidinyl)ethyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.5A resolution. | | Descriptor: | 1-{4-[2-OXO-2-(1-PYRROLIDINYL)ETHYL]PHENYL}-3-( TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX8
 
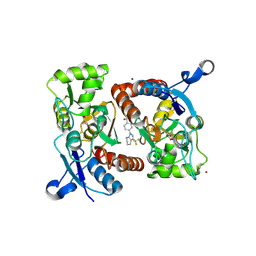 | | Crystal structure of N,N-dimethyl-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)benzamide in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
4P7C
 
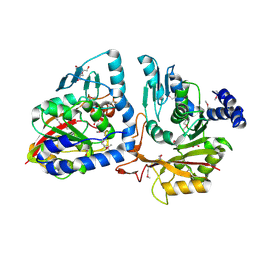 | | Crystal structure of putative methyltransferase from Pseudomonas syringae pv. tomato | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative methyltransferase from Pseudomonas syringae pv. tomato
To Be Published
|
|
4PW0
 
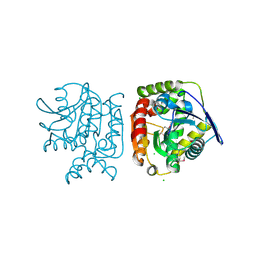 | |
4H0C
 
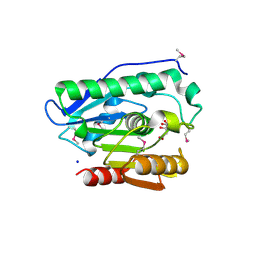 | | Crystal structure of phospholipase/Carboxylesterase from Dyadobacter fermentans DSM 18053 | | Descriptor: | CITRIC ACID, GLYCEROL, Phospholipase/Carboxylesterase, ... | | Authors: | Chang, C, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-07 | | Release date: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of phospholipase/Carboxylesterase from Dyadobacter fermentans DSM 18053
To be Published
|
|
4DIM
 
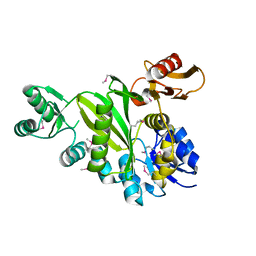 | |
4E9C
 
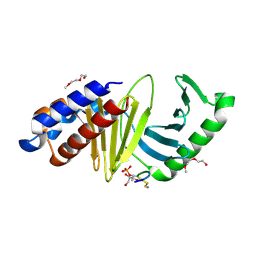 | | The structure of the polo-box domain (PBD) of polo-like kinase 1 (Plk1) in complex with LDPPLHSpTA phosphopeptide | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, LDPPLHSpTA phosphopeptide, ... | | Authors: | Sledz, P, Hyvonen, M, Lang, S, Stubbs, C.J, Abell, C. | | Deposit date: | 2012-03-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-throughput interrogation of ligand binding mode using a fluorescence-based assay.
Angew. Chem. Int. Ed. Engl., 51, 2012
|
|
4MQD
 
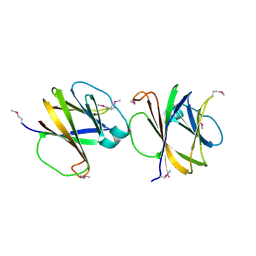 | | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis | | Descriptor: | DNA-entry nuclease inhibitor | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis
To be Published
|
|
2ZC2
 
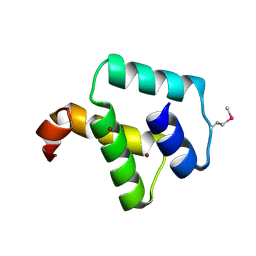 | | Crystal structure of DnaD-like replication protein from Streptococcus mutans UA159, gi 24377835, residues 127-199 | | Descriptor: | DnaD-like replication protein, ZINC ION | | Authors: | Duke, N.E.C, Clancy, S, Duggan, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of DnaD-like replication protein from Streptococcus mutans UA159.
To be Published
|
|
4Q6W
 
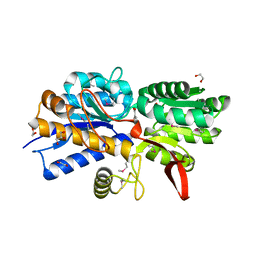 | | Crystal Structure of Periplasmic Binding Protein type 1 from Bordetella pertussis Tohama I complexed with 3-Hydroxy Benzoic Acid | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-23 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Periplasmic Binding Protein type 1 from Bordetella pertussis Tohama I complexed with 3-Hydroxy Benzoic Acid
To be Published, 2014
|
|
5NW7
 
 | | Crystal structure of candida albicans phosphomannose isomerase in complex with inhibitor | | Descriptor: | Mannose-6-phosphate isomerase, ZINC ION, [(2~{R},3~{R},4~{S})-5-diazanyl-2,3,4-tris(oxidanyl)-5-oxidanylidene-pentyl] dihydrogen phosphate | | Authors: | Li de la Sierra-Gallay, I, Ahmad, L, Plancqueel, S, van Tilbeurgh, H, Salmon, L. | | Deposit date: | 2017-05-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of phosphomannose isomerase from Candida albicans complexed with 5-phospho-d-arabinonhydrazide.
FEBS Lett., 592, 2018
|
|
4GYM
 
 | | Crystal structure of Glyoxalase/bleomycin resistance protein/dioxygenase from Conexibacter woesei DSM 14684 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glyoxalase/bleomycin resistance protein/dioxygenase, POTASSIUM ION, ... | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of Glyoxalase/bleomycin resistance protein/dioxygenase from Conexibacter woesei DSM 14684
To be Published
|
|
4HES
 
 | | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula. | | Descriptor: | Beta-lactamase class A-like protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-04 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula.
TO BE PUBLISHED
|
|
