5LMU
 
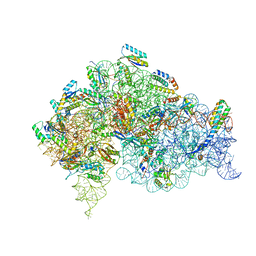 | | Structure of bacterial 30S-IF3-mRNA-tRNA translation pre-initiation complex, closed form (state-4) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMS
 
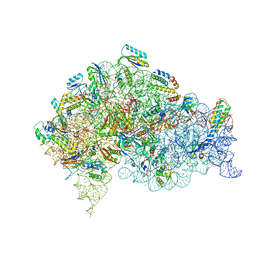 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2C) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5M0Y
 
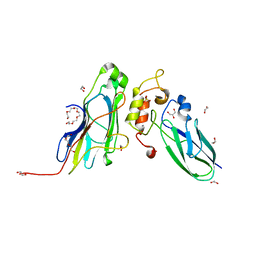 | | Crystal Structure of the CohScaA-XDocCipB type II complex from Clostridium thermocellum at 1.5Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cellulosome anchoring protein cohesin region, ... | | Authors: | Pinheiro, B.A, Bras, J.L, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
7TK0
 
 | |
5LMR
 
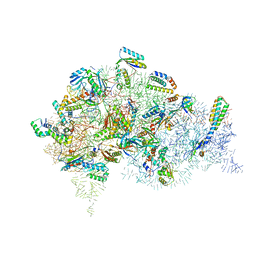 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex(state-2B) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
7TJY
 
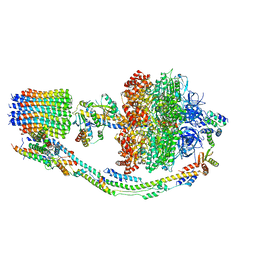 | |
7TJZ
 
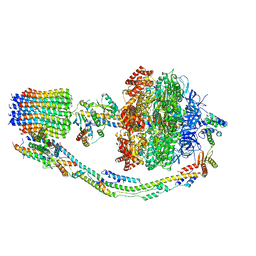 | |
7TK3
 
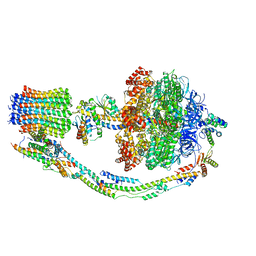 | |
7TK2
 
 | |
7TK4
 
 | |
7TK1
 
 | |
5LMQ
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex, open form (state-2A) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
6I2O
 
 | |
5LMO
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA translation pre-initiation complex (state-1B) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMV
 
 | | Structure of bacterial 30S-IF1-IF2-IF3-mRNA-tRNA translation pre-initiation complex(state-III) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
5LMP
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA translation pre-initiation complex (state-1C) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (5.35 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
6I2T
 
 | | CryoEM reconstruction of full-length, fully-glycosylated human butyrylcholinesterase tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, lamellipodin-derived polyproline peptide | | Authors: | Leung, M.R, van Bezouwen, L.S, Schopfer, L.M, Sussman, J.L, Silman, I, Lockridge, O, Zeev-Ben-Mordehai, T. | | Deposit date: | 2018-11-01 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structure of the native butyrylcholinesterase tetramer reveals a dimer of dimers stabilized by a superhelical assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7T1D
 
 | | Human SIRT2 in complex with small molecule 359 | | Descriptor: | 1,2-ETHANEDIOL, 7-(2,4-dimethyl-1H-imidazol-1-yl)-2-(5-{[4-(1H-pyrazol-1-yl)phenyl]methyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline, DIMETHYL SULFOXIDE, ... | | Authors: | Kulp, J.L, Remiszewski, S, Todd, M, Chiang, L.W. | | Deposit date: | 2021-12-01 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An allosteric inhibitor of sirtuin 2 deacetylase activity exhibits broad-spectrum antiviral activity.
J.Clin.Invest., 133, 2023
|
|
6X5C
 
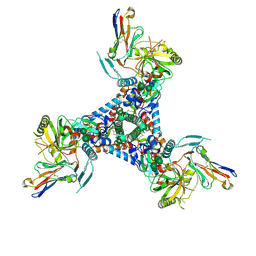 | | Asymmetric model of CD4-bound B41 HIV-1 Env SOSIP in complex with small molecule GO52 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2020-05-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6X5B
 
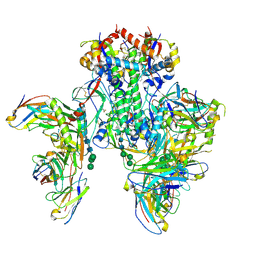 | | Symmetric model of CD4- and 17-bound B41 HIV-1 Env SOSIP in complex with small molecule GO52 | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2020-05-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
6XYS
 
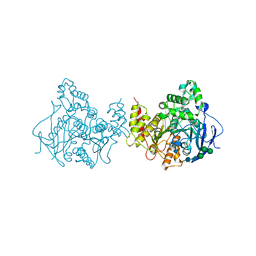 | | Update of native acetylcholinesterase from Drosophila Melanogaster | | Descriptor: | ACETATE ION, Acetylcholinesterase, CHLORIDE ION, ... | | Authors: | Nachon, F, Sussman, J.L. | | Deposit date: | 2020-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A Second Look at the Crystal Structures ofDrosophila melanogasterAcetylcholinesterase in Complex with Tacrine Derivatives Provides Insights Concerning Catalytic Intermediates and the Design of Specific Insecticides.
Molecules, 25, 2020
|
|
6XOD
 
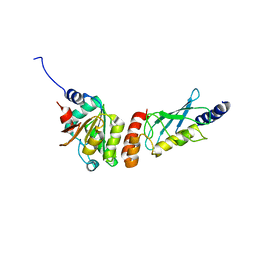 | | Crystal structure of the PEX4-PEX22 protein complex from Arabidopsis thaliana | | Descriptor: | Peroxisome biogenesis protein 22, Protein PEROXIN-4 | | Authors: | Olmos Jr, J.L, Bradford, S.E, Miller, M.D, Xu, W, Wright, Z.J, Bartel, B, Phillips Jr, G.N. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane
Front Cell Dev Biol, 10, 2022
|
|
5G5D
 
 | | Crystal Structure of the CohScaC2-XDocCipA type II complex from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CELLULOSOMAL-SCAFFOLDING PROTEIN A, CELLULOSOME ANCHORING PROTEIN COHESIN REGION | | Authors: | Carvalho, A.L, A Bras, J.L, Najmudin, S.H, Pinheiro, B.A, Fontes, C.M.G.A. | | Deposit date: | 2016-05-23 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
5GKP
 
 | |
5GRM
 
 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
