2KNR
 
 | | Solution structure of protein Atu0922 from A. tumefaciens. Northeast Structural Genomics Consortium target AtT13. Ontario Center for Structural Proteomics target ATC0905 | | Descriptor: | Uncharacterized protein ATC0905 | | Authors: | Gutmanas, A, Yee, S, Lemak, A, Fares, A, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein atc0905 from A. tumefaciens
To be Published
|
|
2KDB
 
 | | Solution Structure of human ubiquitin-like domain of Herpud2_9_85, Northeast Structural Genomics Consortium (NESG) target HT53A | | Descriptor: | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 2 protein | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Gutmanas, A, Doherty, R, Semesi, A, Dhe-Paganon, S, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-06 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of human homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 2 (Herpud2 or Herp)
To be Published
|
|
8ZMF
 
 | | Crystal structure of an inverse agonist antipsychotic drug derivative-bound 5-HT2C | | Descriptor: | 1-[(4-fluorophenyl)methyl]-1-[(8~{S})-5-methyl-5-azaspiro[2.5]octan-8-yl]-3-[[4-(2-methylpropoxy)phenyl]methyl]urea, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562 | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
8ZMG
 
 | | Crystal structure of an inverse agonist antipsychotic drug pimavanserin-bound 5-HT2A | | Descriptor: | 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, Pimavanserin | | Authors: | Oguma, T, Asada, H, Sekiguchi, Y, Imono, M, Iwata, S, Kusakabe, K. | | Deposit date: | 2024-05-23 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Dual 5-HT 2A and 5-HT 2C Receptor Inverse Agonist That Affords In Vivo Antipsychotic Efficacy with Minimal hERG Inhibition for the Treatment of Dementia-Related Psychosis.
J.Med.Chem., 67, 2024
|
|
6LX2
 
 | | Potato D-enzyme complexed with CA26 | | Descriptor: | 4-alpha-glucanotransferase, chloroplastic/amyloplastic, 4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose-(1-4)-4-deoxy-alpha-D-glucopyranose, ... | | Authors: | Unno, H, Imamura, K. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
6LX1
 
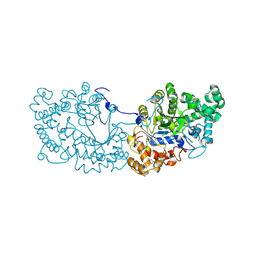 | | Potato D-enzyme complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4-alpha-glucanotransferase, chloroplastic/amyloplastic, ... | | Authors: | Unno, H, Imamura, K. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural analysis and reaction mechanism of the disproportionating enzyme (D-enzyme) from potato.
Protein Sci., 29, 2020
|
|
4Y16
 
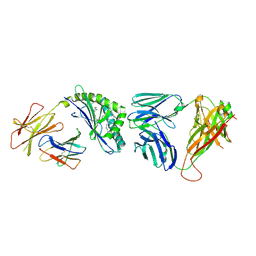 | | Crystal structure of the mCD1d/NC-aGC/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Nemcovic, M. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lipid and Carbohydrate Modifications of alpha-Galactosylceramide Differently Influence Mouse and Human Type I Natural Killer T Cell Activation.
J.Biol.Chem., 290, 2015
|
|
2JOQ
 
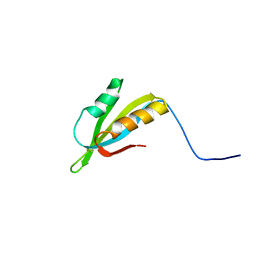 | | Solution Structure of Protein HP0495 from H. pylori; Northeast structural genomics consortium target PT2; Ontario Centre for Structural Proteomics target HP0488 | | Descriptor: | Hypothetical protein HP_0495 | | Authors: | Gutmanas, A, Lemak, A, Yee, A, Karra, M, Srisailam, S, Semesi, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Protein HP0495 from H. pylori
To be Published
|
|
7DBJ
 
 | | Crystal structure of human LDHB in complex with NADH, oxamate, and AXKO-0046 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase B chain, ... | | Authors: | Sogabe, S, Miwa, M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Identification of the first highly selective inhibitor of human lactate dehydrogenase B.
Sci Rep, 11, 2021
|
|
7DBK
 
 | | Crystal structure of human LDHB in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase B chain | | Authors: | Sogabe, S, Miwa, M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Identification of the first highly selective inhibitor of human lactate dehydrogenase B.
Sci Rep, 11, 2021
|
|
2JQ4
 
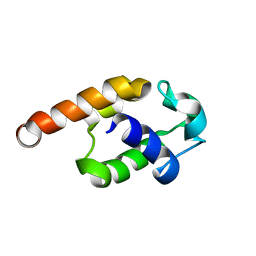 | | Complete resonance assignments and solution structure calculation of ATC2521 (NESG ID: AtT6) from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu2571 | | Authors: | Srisailam, S, Lemak, A, Yee, A, Karra, M.D, Lukin, J.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a protein (ATC2521)of unknown function from Agrobacterium tumefaciens
To be Published
|
|
2JN4
 
 | | Solution NMR Structure of Protein RP4601 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium Target RpT2; Ontario Center for Structural Proteomics Target RP4601. | | Descriptor: | Hypothetical protein fixU, nifT | | Authors: | Lemak, A, Srisailam, S, Yee, A, Karra, M.D, Lukin, J.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-22 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hypothetical protein RP4601 from Rhodopseudomonas palustris
To be Published
|
|
2KEO
 
 | | Solution NMR structure of human protein HS00059, cytochrome-b5-like domain of the HERC2 E3 ligase. Northeast structural genomics consortium (NESG) target ht98a | | Descriptor: | Probable E3 ubiquitin-protein ligase HERC2 | | Authors: | Lemak, A, Gutmanas, A, Fares, C, Quyang, H, Li, Y, Montelione, G, Arrowsmith, C, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of human protein HS00059
To be Published
|
|
2JTV
 
 | | Solution Structure of protein RPA3401, Northeast Structural Genomics Consortium Target RpT7, Ontario Center for Structural Proteomics Target RP3384 | | Descriptor: | Protein of Unknown Function | | Authors: | Ignatchenko, A, Gutmanas, A, Lemak, A, Yee, A, Karra, M, Srisailam, S, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of protein RPA3401, Northeast Structural Genomics Consortium Target RpT7, Ontario Center for Structural Proteomics Target RP3384
TO BE PUBLISHED
|
|
2KKU
 
 | | Solution structure of protein af2351 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium target AtT9/Ontario Center for Structural Proteomics Target af2351 | | Descriptor: | Uncharacterized protein | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Rumpel, S, Semest, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein af2351 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium target AtT9/Ontario Center for Structural Proteomics Target af2351
To be Published
|
|
2KR1
 
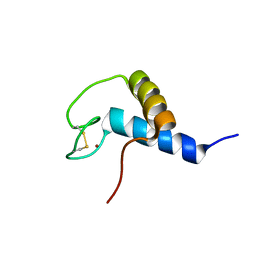 | | Solution NMR structure of zinc binding N-terminal domain of ubiquitin-protein ligase E3A from Homo Sapiens. Northeast Structural Genomics Consortium (NESG) target HR3662 | | Descriptor: | Ubiquitin protein ligase E3A, ZINC ION | | Authors: | Lemak, A, Yee, A, Fares, C, Semesi, A, Xiao, R, Montelione, G, Dhe-Paganon, S, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Zn-binding AZUL domain of human ubiquitin protein ligase Ube3A.
J.Biomol.Nmr, 51, 2011
|
|
3WJ1
 
 | | Crystal structure of SSHESTI | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
3WNK
 
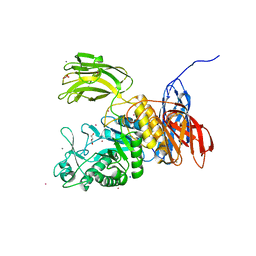 | | Crystal Structure of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | ACETATE ION, CADMIUM ION, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
4QN0
 
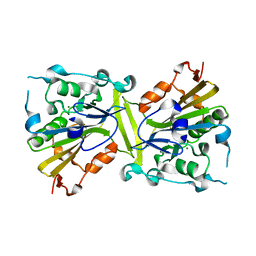 | | Crystal structure of the CPS-6 mutant Q130K | | Descriptor: | Endonuclease G, mitochondrial, MAGNESIUM ION | | Authors: | Lin, J.L.J, Yuan, H.S. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Oxidative Stress Impairs Cell Death by Repressing the Nuclease Activity of Mitochondrial Endonuclease G
Cell Rep, 16, 2016
|
|
4HL0
 
 | |
2E0T
 
 | | Crystal structure of catalytic domain of dual specificity phosphatase 26, MS0830 from Homo sapiens | | Descriptor: | Dual specificity phosphatase 26 | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Chen, L, Liu, Z.J, Wang, B.C, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-resolution crystal structure of the catalytic domain of human dual-specificity phosphatase 26.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4RWZ
 
 | | Crystal structure of the antibiotic-resistance methyltransferase Kmr | | Descriptor: | Putative rRNA methyltransferase | | Authors: | Savic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | 30S Subunit-Dependent Activation of the Sorangium cellulosum So ce56 Aminoglycoside Resistance-Conferring 16S rRNA Methyltransferase Kmr.
Antimicrob.Agents Chemother., 59, 2015
|
|
4RX1
 
 | | Crystal Structure of antibiotic-resistance methyltransferase Kmr | | Descriptor: | GLYCEROL, IODIDE ION, Putative rRNA methyltransferase | | Authors: | Savic, M. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | 30S Subunit-Dependent Activation of the Sorangium cellulosum So ce56 Aminoglycoside Resistance-Conferring 16S rRNA Methyltransferase Kmr.
Antimicrob.Agents Chemother., 59, 2015
|
|
1VF7
 
 | | Crystal structure of the membrane fusion protein, MexA of the multidrug transporter | | Descriptor: | Multidrug resistance protein mexA | | Authors: | Akama, H, Matsuura, T, Kashiwagi, S, Yoneyama, H, Tsukihara, T, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-04-09 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa
J.Biol.Chem., 279, 2004
|
|
4TXW
 
 | | Crystal structure of CBM32-4 from the Clostridium perfringens NagH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-07-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution Structure and Dynamics of Full-length GH84A, a multimodular B-N-acetylglucosaminidase from Clostridium perfringens
To Be Published
|
|
