8WBD
 
 | |
8WBG
 
 | |
8WBF
 
 | |
8WBH
 
 | |
8ZE3
 
 | |
8ZE2
 
 | |
8ZE0
 
 | |
8ZDZ
 
 | |
5JHG
 
 | | Crystal structure of the complex between the human RhoA and the DH/PH domain of human ARHGEF11 | | Descriptor: | GLYCEROL, Rho guanine nucleotide exchange factor 11, Transforming protein RhoA | | Authors: | Wang, R, Chen, Q, Zhang, H, Yan, Z, Li, J, Miao, L, Wang, F. | | Deposit date: | 2016-04-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA bound with its inhibitor and ARHGEF11
To Be Published
|
|
8G1P
 
 | | Co-crystal structure of Compound 11 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Ghimire Rijal, S, Wurz, R.P, Vaish, A. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation.
Nat Commun, 14, 2023
|
|
8G1Q
 
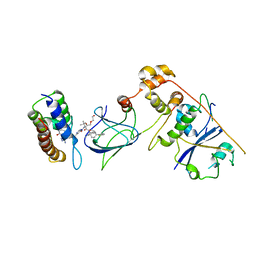 | | Co-crystal structure of Compound 1 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongin-B, Elongin-C, ... | | Authors: | Ghimire Rijal, S, Wurz, R.P, Vaish, A. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation.
Nat Commun, 14, 2023
|
|
7V4X
 
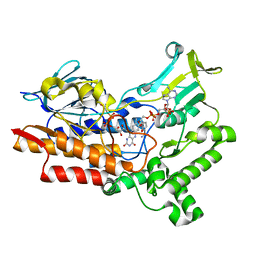 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V50
 
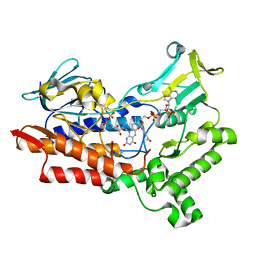 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V51
 
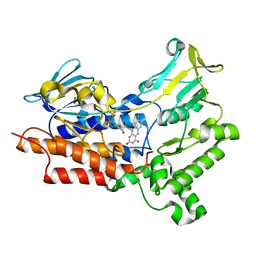 | | BVMO_negative mutant D432V | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H.-L. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
8ZNZ
 
 | | CD73 bound with HB0045 | | Descriptor: | 5'-nucleotidase, CALCIUM ION, HB0038 Fab heavy chain, ... | | Authors: | Xu, J, Wan, L, He, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | An antibody cocktail targeting two different CD73 epitopes enhances enzyme inhibition and tumor control.
Nat Commun, 15, 2024
|
|
8I5M
 
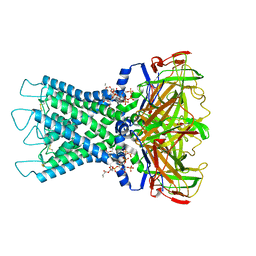 | | Rat Kir4.1 in complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
8I5N
 
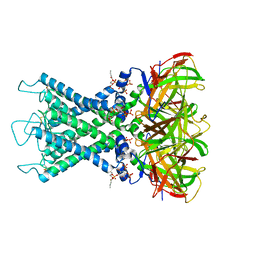 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
6M3M
 
 | |
6IV8
 
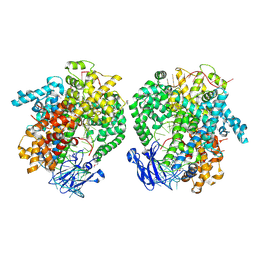 | | the selenomethionine(SeMet)-derived Cas13d binary complex | | Descriptor: | MAGNESIUM ION, RNA (51-MER), RNA (53-MER), ... | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
8J1I
 
 | | Crystal Structure of EphA8/SASH1 Complex | | Descriptor: | Ephrin type-A receptor 8, SAM and SH3 domain-containing protein 1 | | Authors: | Liu, W, Li, J, Ding, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | SASH1: A Novel Eph Receptor Partner and Insights into SAM-SAM Interactions.
J.Mol.Biol., 435, 2023
|
|
7VFX
 
 | | The structure of Formyl Peptide Receptor 1 in complex with Gi and peptide agonist fMIFL | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X.K, Chen, G, Liao, Q.W, Du, Y, Hu, H.L, Ye, D.Q. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for recognition of N-formyl peptides as pathogen-associated molecular patterns.
Nat Commun, 13, 2022
|
|
8K7O
 
 | | De novo design protein -T03 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8I
 
 | | De novo design protein -N14 | | Descriptor: | De novo design protein -N14 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-30 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K8F
 
 | | De novo design protein -N7 | | Descriptor: | De novo design protein N7, GLYCEROL | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
8K7Z
 
 | | De novo design protein -N1 | | Descriptor: | De novo design protein | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-07-27 | | Release date: | 2024-07-31 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
