5VS5
 
 | | ABA-mimicking ligand AMF2alpha in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(2,3-difluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-W, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
5VSQ
 
 | | ABA-mimicking ligand AMF2beta in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(3,5-difluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.618 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
5VSR
 
 | | ABA-mimicking ligand AMF4 in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-(2,3,5,6-tetrafluoro-4-methylphenyl)methanesulfonamide, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.618 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
5VRO
 
 | | ABA-mimicking ligand AMF1beta in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(3-fluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
8INQ
 
 | |
8INT
 
 | |
8INW
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) K90R Mutant in Complex with Inhibitor nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
5VT7
 
 | | ABA-mimicking ligand AMC1beta in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | Deposit date: | 2017-05-15 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.624 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
8INY
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) K90R Mutant in Complex with Inhibitor ensitrelvir | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
8INU
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
8INX
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor ensitrelvir | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
5ZQY
 
 | | Crystal structure of a poly(ADP-ribose) glycohydrolase | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, M, Yuan, Z, Ma, Y, Wang, J, Liu, X. | | Deposit date: | 2018-04-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.577 Å) | | Cite: | Structure-function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation.
J. Biol. Chem., 293, 2018
|
|
8VVY
 
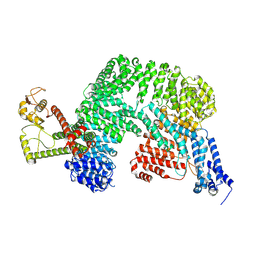 | | Human Cullin-1 in complex with CAND2 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 2 | | Authors: | Kenny, S, Liu, X, Das, C. | | Deposit date: | 2024-01-31 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Cryo-EM structure of Human CAND2 in complex with Cullin-1
To Be Published
|
|
5VR7
 
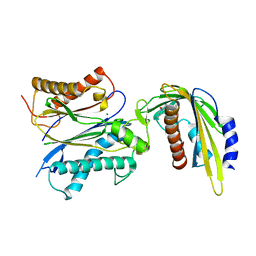 | | ABA-mimicking ligand AMF1alpha in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(2-fluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
9G0I
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the noncovalently bound inhibitor C5N17B | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, IMIDAZOLE, ... | | Authors: | Yang, C.-C, Strater, N, Sylvester, K, Muller, C.E, Yang, M, Lee, M.K, Gao, S, Song, L, Liu, X, Kim, M, Zhan, P. | | Deposit date: | 2024-07-08 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Miniaturized Modular Click Chemistry-enabled Rapid Discovery of Unique SARS-CoV-2 M pro Inhibitors With Robust Potency and Drug-like Profile.
Adv Sci, 11, 2024
|
|
9G0H
 
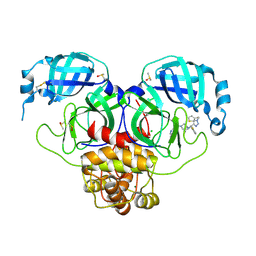 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the noncovalently bound inhibitor C5N17A | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, C.-C, Strater, N, Sylvester, K, Muller, C.E, Yang, M, Lee, M.K, Gao, S, Song, L, Liu, X, Kim, M, Zhan, P. | | Deposit date: | 2024-07-08 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Miniaturized Modular Click Chemistry-enabled Rapid Discovery of Unique SARS-CoV-2 M pro Inhibitors With Robust Potency and Drug-like Profile.
Adv Sci, 11, 2024
|
|
9IVM
 
 | | Cryo-EM structure of the GLP-1(9-36)-bound human GLP-1R-Gs complex in the presence of LSN3318839 | | Descriptor: | (2~{R})-2-[2-[1-[(1~{R})-1-[2,6-bis(chloranyl)-3-cyclopropyl-phenyl]ethyl]imidazo[4,5-c]pyridin-6-yl]phenyl]propanoic acid, GLP-1(9-36), Glucagon-like peptide 1 receptor, ... | | Authors: | Li, J, Li, G, Mai, Y, Liu, X, Yang, D, Zhou, Q, Wang, M.-W. | | Deposit date: | 2024-07-24 | | Release date: | 2024-11-13 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular basis of enhanced GLP-1 signaling mediated by GLP-1(9-36) in conjunction with LSN3318839.
Acta Pharm Sin B, 14, 2024
|
|
9IVG
 
 | | Cryo-EM structure of the GLP-1(9-36)-bound human GLP-1R-Gs complex | | Descriptor: | GLP-1(9-36), Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, J, Li, G, Mai, Y, Liu, X, Yang, D, Zhou, Q, Wang, M.-W. | | Deposit date: | 2024-07-23 | | Release date: | 2024-11-27 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of enhanced GLP-1 signaling mediated by GLP-1(9-36) in conjunction with LSN3318839.
Acta Pharm Sin B, 14, 2024
|
|
8JWE
 
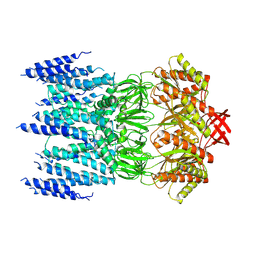 | |
9J14
 
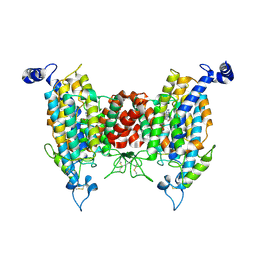 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state at pH 5.5 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state at pH 5.5
To Be Published
|
|
9J16
 
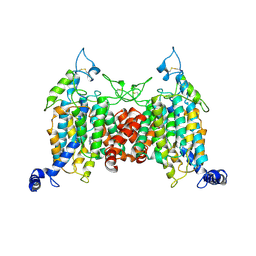 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the adenine-bound state at pH 7.4 | | Descriptor: | ADENINE, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the adenine-bound state at pH 7.4
To Be Published
|
|
9J18
 
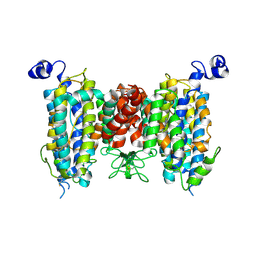 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-2 at pH 7.4 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-2 at pH 7.4
To Be Published
|
|
9J17
 
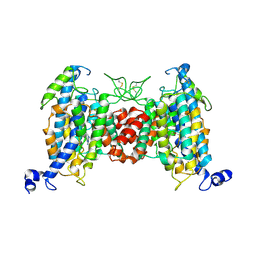 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-1 at pH 7.4 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the trans-Zeatin-bound state-1 at pH 7.4
To Be Published
|
|
9J15
 
 | | Structure of the wild-type AZG2 in Arabidopsis thaliana in the adenine-bound state at pH 5.5 | | Descriptor: | ADENINE, Adenine/guanine permease AZG2 | | Authors: | Sun, L, Liu, X, Wei, H, Yang, Z, Ying, W. | | Deposit date: | 2024-08-03 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the wild-type AZG2 in Arabidopsis thaliana in the adenine-bound state at pH 5.5
To Be Published
|
|
9J12
 
 | |
