7BTA
 
 | | Crystal structure of Rheb D60K mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-03-31 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTC
 
 | | Crystal structure of Rheb Y35N mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTD
 
 | | Crystal structure of Rheb Y35N mutant bound to GppNHp | | Descriptor: | GTP-binding protein Rheb, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
8J8D
 
 | | Crystal structure of SRCR domain 11 of DMBT1 | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein | | Authors: | Zhang, C, Lu, P, Nagata, K. | | Deposit date: | 2023-05-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Refolding, Crystallization, and Crystal Structure Analysis of a Scavenger Receptor Cysteine-Rich Domain of Human Salivary Agglutinin Expressed in Escherichia coli.
Protein J., 43, 2024
|
|
8WZS
 
 | |
7E6V
 
 | | The crystal structure of foot-and-mouth disease virus(FMDV) 2C protein 97-318aa | | Descriptor: | ACETATE ION, Protein 2C | | Authors: | Zhang, C, Wojdyla, J.A, Qin, B, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2021-02-24 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | An anti-picornaviral strategy based on the crystal structure of foot-and-mouth disease virus 2C protein.
Cell Rep, 40, 2022
|
|
8J8T
 
 | |
6L7O
 
 | | cryo-EM structure of cyanobacteria Fd-NDH-1L complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
8YRE
 
 | | Crystal structure of Arabidopsis VTC1-KJC2 | | Descriptor: | ADP-glucose pyrophosphorylase family protein, Mannose-1-phosphate guanylyltransferase 1, SULFATE ION | | Authors: | Zhang, C, Liu, L. | | Deposit date: | 2024-03-21 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structural insight into the regulation of GDP-mannose pyrophosphorylase activity of Arabidopsis KJC2-VTC1 complex
To Be Published
|
|
7BWQ
 
 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
7BYY
 
 | | Crystal structure of bacterial toxin | | Descriptor: | Acetyltransferase | | Authors: | Zhang, C, Yashiro, Y, Tomita, K. | | Deposit date: | 2020-04-25 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Substrate specificities of Escherichia coli ItaT that acetylates aminoacyl-tRNAs.
Nucleic Acids Res., 48, 2020
|
|
4XV2
 
 | |
4XV3
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7922 | | Descriptor: | N'-{3-[5-(2-aminopyrimidin-4-yl)-2-tert-butyl-1,3-thiazol-4-yl]-2-fluorophenyl}-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV9
 
 | | B-Raf Kinase domain in complex with PLX5568 | | Descriptor: | N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}-4-(trifluoromethyl)benzenesulfonamide, SULFATE ION, Serine/threonine-protein kinase B-raf | | Authors: | zhang, Y, zhang, c, wang, w. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4XV1
 
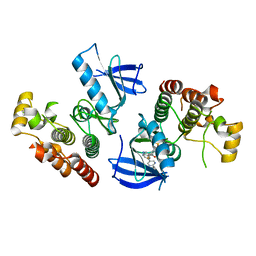 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7904 | | Descriptor: | N'-(3-{[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2015-01-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
4HW7
 
 | |
4HVS
 
 | |
4RL5
 
 | |
6EOA
 
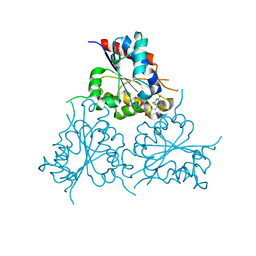 | | Crystal Structure of HAL3 from Cryptococcus neoformans | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phosphopantothenoylcysteine decarboxylase | | Authors: | Reverter, D, Zhang, C, Molero, C, Arino, J. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Characterization of the atypical Ppz/Hal3 phosphatase system from the pathogenic fungus Cryptococcus neoformans.
Mol.Microbiol., 111, 2019
|
|
8IQY
 
 | |
8IQV
 
 | |
8IQW
 
 | |
8IQZ
 
 | |
5Z9J
 
 | | Identification of the functions of unusual cytochrome p450-like monooxygenases involved in microbial secondary metablism | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like enzyme, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lu, M, Lin, L, Zhang, C, Chen, Y. | | Deposit date: | 2018-02-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Riboflavin Is Directly Involved in the N-Dealkylation Catalyzed by Bacterial Cytochrome P450 Monooxygenases.
Chembiochem, 2020
|
|
5Z9I
 
 | |
