3J9D
 
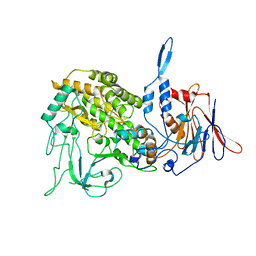 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | Outer capsid protein VP2, ZINC ION | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3J9E
 
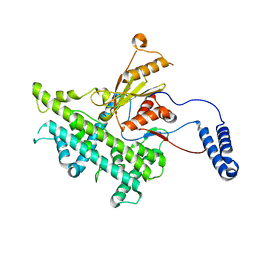 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | VP5 | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3JB7
 
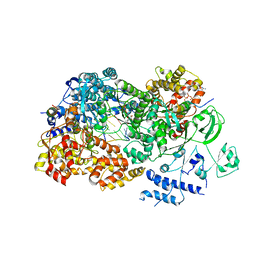 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
2RFD
 
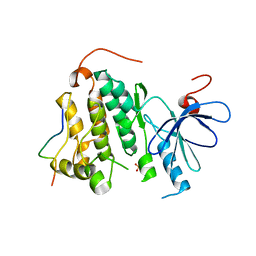 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor, SULFATE ION | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RF9
 
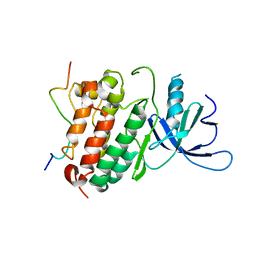 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RFE
 
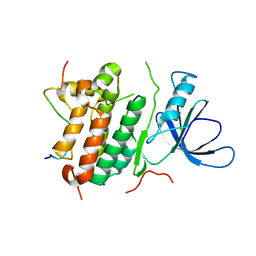 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
1H0C
 
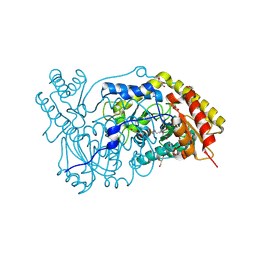 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
5K3G
 
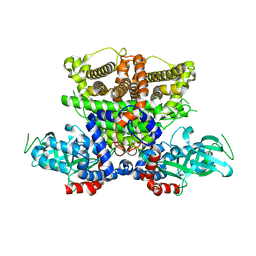 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-I | | Descriptor: | Acyl-coenzyme A oxidase | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3J
 
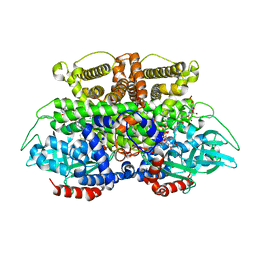 | | Crystals structure of Acyl-CoA oxidase-2 in Caenorhabditis elegans bound with FAD, ascaroside-CoA, and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1HQK
 
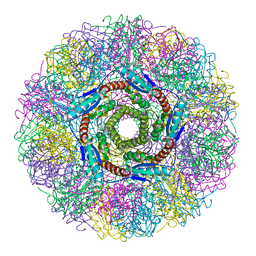 | | CRYSTAL STRUCTURE ANALYSIS OF LUMAZINE SYNTHASE FROM AQUIFEX AEOLICUS | | Descriptor: | 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE | | Authors: | Zhang, X, Meining, W, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons.
J.Mol.Biol., 306, 2001
|
|
9NS9
 
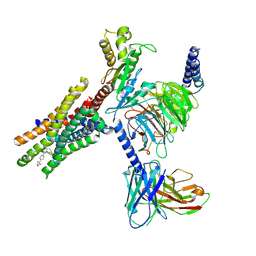 | | Cryo-EM structure of Gi-coupled FFA2 in complex with TUG-1375 and compound 187 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, 4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-7-(2-fluorobenzene-1-sulfonyl)-2-methyl-5H-pyrrolo[3,2-d]pyrimidin-6-amine, Free fatty acid receptor 2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2025-03-16 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric modulation and biased signalling at free fatty acid receptor 2.
Nature, 2025
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
5YB2
 
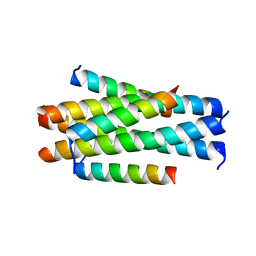 | | Crystal structure of LP-11/N44 | | Descriptor: | Envelope glycoprotein, LP-11 | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
5YB4
 
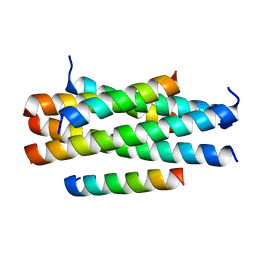 | | Crystal structure of HP23LN36KR | | Descriptor: | HP23L, N36KR | | Authors: | Zhang, X, Wang, X, He, Y. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Mechanisms of Action of Short-Peptide HIV-1 Fusion Inhibitors Targeting the Gp41 Pocket
Front Cell Infect Microbiol, 8, 2018
|
|
6IMB
 
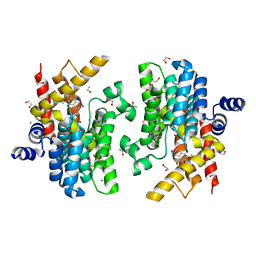 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMI
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-ethoxy-7-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMD
 
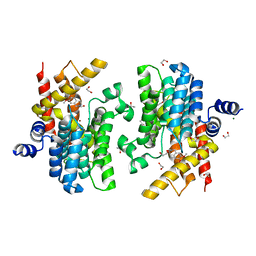 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2010-04-14 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
5KQA
 
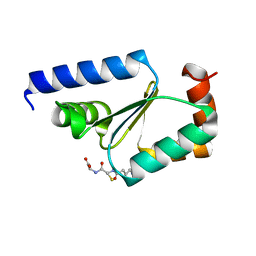 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | Descriptor: | GLUTATHIONE, Glutaredoxin-glutathione complex | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
1NCN
 
 | |
3J27
 
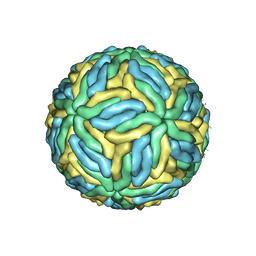 | | CryoEM structure of Dengue virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Zhang, X, Ge, P, Yu, X, Brannan, J.M, Bi, G, Zhang, Q, Schein, S, Zhou, Z.H. | | Deposit date: | 2012-09-26 | | Release date: | 2012-12-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the mature dengue virus at 3.5-A resolution.
Nat.Struct.Mol.Biol., 20, 2012
|
|
2Y3A
 
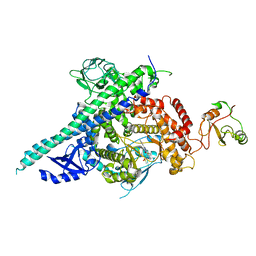 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | Authors: | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
1F3L
 
 | |
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | Descriptor: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
1ML9
 
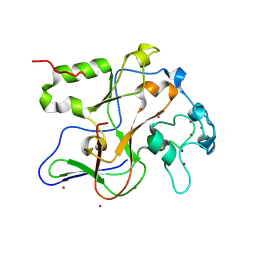 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | Descriptor: | Histone H3 methyltransferase DIM-5, UNKNOWN LIGAND, ZINC ION | | Authors: | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | Deposit date: | 2002-08-30 | | Release date: | 2002-10-23 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
