7DDC
 
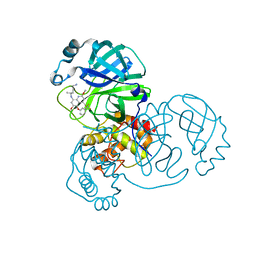 | | Crystal structure of SARS-CoV-2 main protease in complex with Tafenoquine | | Descriptor: | 3C-like proteinase, Tafenoquine | | Authors: | Chen, Y, Wang, Y.C, Yang, C.S, Hung, M.C. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Tafenoquine and its derivatives as inhibitors for the severe acute respiratory syndrome coronavirus 2.
J.Biol.Chem., 298, 2022
|
|
4RZF
 
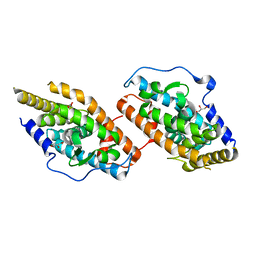 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
4RZG
 
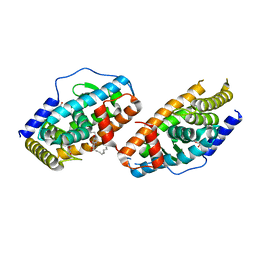 | | Crystal Structure Analysis of the DNPA-bounded NUR77 Ligand binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, pentyl (3,5-dihydroxy-2-nonanoylphenyl)acetate | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
4HZ9
 
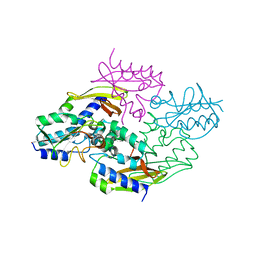 | | Crystal structure of the type VI native effector-immunity complex Tae3-Tai3 from Ralstonia pickettii | | Descriptor: | Putative cytoplasmic protein, Putative periplasmic protein | | Authors: | Dong, C, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2012-11-14 | | Release date: | 2013-08-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3
Biochem.J., 454, 2013
|
|
4HZB
 
 | | Crystal structure of the type VI SeMet effector-immunity complex Tae3-Tai3 from Ralstonia pickettii | | Descriptor: | Putative cytoplasmic protein, Putative periplasmic protein | | Authors: | Dong, C, Zhang, H, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2012-11-15 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the inhibition of type VI effector Tae3 by its immunity protein Tai3
Biochem.J., 454, 2013
|
|
4E8B
 
 | |
4EQ6
 
 | |
4HFK
 
 | |
4HFF
 
 | |
4HFL
 
 | |
3TKA
 
 | | crystal structure and solution saxs of methyltransferase rsmh from E.coli | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Ribosomal RNA small subunit methyltransferase H, S-ADENOSYLMETHIONINE, ... | | Authors: | Gao, Z.Q, Wei, Y, Zhang, H, Dong, Y.H. | | Deposit date: | 2011-08-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA.
J.Struct.Biol., 179, 2012
|
|
