7NNK
 
 | | Crystal structure of S116A mutant of hydroxy ketone aldolase (SwHKA) from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, HpcH/HpaI aldolase, MAGNESIUM ION | | Authors: | Justo, I, Marsden, S.R, Hanefeld, U, Bento, I. | | Deposit date: | 2021-02-25 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Induced Movement of the Metal Cofactor between Active and Resting State.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4IWT
 
 | |
4IVV
 
 | |
5N7E
 
 | | Crystal structure of the Dbl-homology domain of Bcr-Abl in complex with monobody Mb(Bcr-DH_4). | | Descriptor: | Breakpoint cluster region protein, Mb(Bcr-DH_4) | | Authors: | Reckel, S, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2017-02-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
5N6R
 
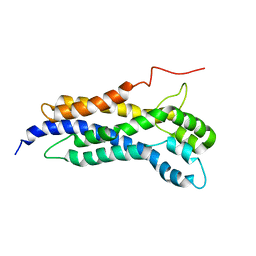 | | Solution structure of the Dbl-homology domain of Bcr-Abl | | Descriptor: | Breakpoint cluster region protein | | Authors: | Reckel, S, Lohr, F, Buchner, L, Guntert, P, Dotsch, V, Hantschel, O. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
5OC7
 
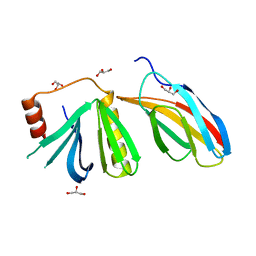 | | Crystal structure of the pleckstrin-homology domain of Bcr-Abl in complex with monobody Mb(Bcr-PH_4). | | Descriptor: | Breakpoint cluster region protein,pleckstrin-homology domain of Bcr-Abl, D-MYO-INOSITOL-4,5-BISPHOSPHATE, GLYCEROL, ... | | Authors: | Reckel, S, Reynaud, A, Pojer, F, Hantschel, O. | | Deposit date: | 2017-06-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
2BWC
 
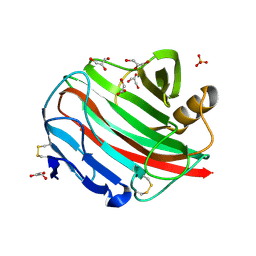 | |
2BWA
 
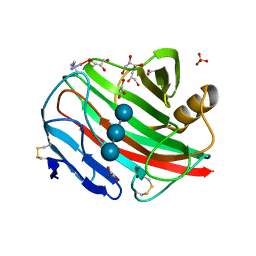 | |
2BW8
 
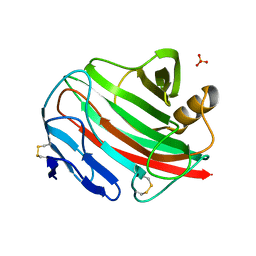 | |
4EY0
 
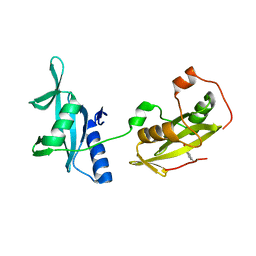 | | Structure of tandem SH2 domains from PLCgamma1 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
7A25
 
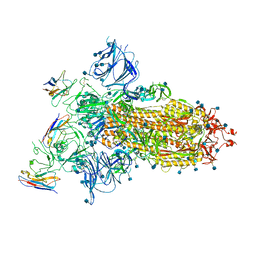 | |
4FBN
 
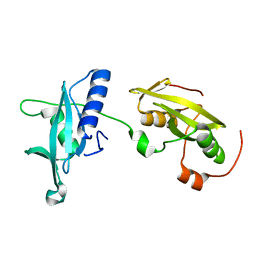 | | Insights into structural integration of the PLCgamma regulatory region and mechanism of autoinhibition and activation based on key roles of SH2 domains | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | | Authors: | Cole, A.R, Mas-Droux, C.P, Bunney, T.D, Katan, M. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Integration of the PLCgamma Interaction Domains Critical for Regulatory Mechanisms and Signaling Deregulation.
Structure, 20, 2012
|
|
5M89
 
 | | Spliceosome component | | Descriptor: | Spliceosome WD40 Sc | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
5M88
 
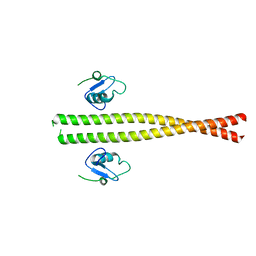 | | Spliceosome component | | Descriptor: | Core | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
6RHV
 
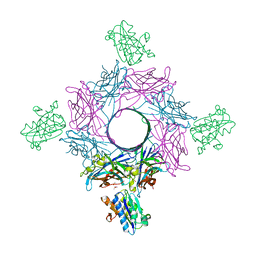 | | Crystal structure of mouse CD11b I-domain (CD11b-I) in complex with Staphylococcus aureus octameric bi-component leukocidin LukGH (LukH K319A mutant) | | Descriptor: | Beta-channel forming cytolysin, DIMETHYL SULFOXIDE, Integrin alpha-M, ... | | Authors: | Trstenjak, N, Milic, D, Djinovic-Carugo, K, Badarau, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5M8C
 
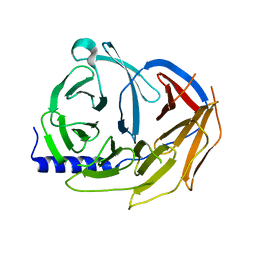 | | Spliceosome component | | Descriptor: | Pre-mRNA-processing factor 19 | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
6RHW
 
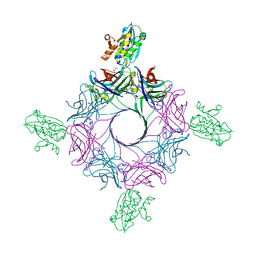 | | Crystal structure of human CD11b I-domain (CD11b-I) in complex with Staphylococcus aureus octameric bi-component leukocidin LukGH | | Descriptor: | Beta-channel forming cytolysin, DIMETHYL SULFOXIDE, Integrin alpha-M, ... | | Authors: | Trstenjak, N, Milic, D, Djinovic-Carugo, K, Badarau, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3QS9
 
 | |
3QS7
 
 | |
6HXL
 
 | |
6HXJ
 
 | |
6HXQ
 
 | |
6HXK
 
 | |
6HXM
 
 | |
6HXO
 
 | |
