7QXQ
 
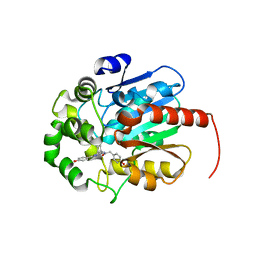 | | Coelenteramide-bound Renilla-type luciferase (AncFT) | | Descriptor: | Fragment transplantation onto hyperstable ancestor of haloalkane dehalogenases and Renilla luciferase (Anc-FT), N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE | | Authors: | Marek, M, Schenkmayerova, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
8AX8
 
 | |
8AX9
 
 | |
6TY7
 
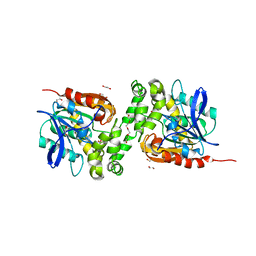 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
6S97
 
 | |
4F60
 
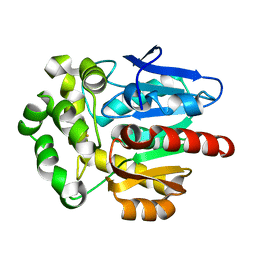 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (T148L, G171Q, A172V, C176F). | | Descriptor: | FLUORIDE ION, Haloalkane dehalogenase | | Authors: | Plevaka, M, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4F5Z
 
 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (L95V, A172V). | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Kulik, D, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4E46
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA in complex with 2-propanol | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Stsiapanava, A, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Expansion of access tunnels and active-site cavities influence activity of haloalkane dehalogenases in organic cosolvents.
Chembiochem, 14, 2013
|
|
8CLN
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus, SeMet derivative for SAD phasing | | Descriptor: | Hydrolase | | Authors: | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
8CLO
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase | | Authors: | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
8CLV
 
 | |
8CLQ
 
 | |
8OE6
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
8OE2
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA223 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
1K5P
 
 | | Hydrolytic haloalkane dehalogenase LINB from sphingomonas paucimobilis UT26 at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1K63
 
 | | Complex of hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis with UT26 2-BROMO-2-PROPENE-1-OL at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, 2-BROMO-2-PROPENE-1-OL, BROMIDE ION, ... | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-15 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1IZ8
 
 | | Re-refinement of the structure of hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Streltsov, V.A. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1IZ7
 
 | |
1MJ5
 
 | | LINB (haloalkane dehalogenase) from sphingomonas paucimobilis UT26 at atomic resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Oakley, A.J, Damborsky, J, Wilce, M.C. | | Deposit date: | 2002-08-27 | | Release date: | 2003-08-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 A resolution: dynamics of catalytic residues.
Biochemistry, 43, 2004
|
|
8AQH
 
 | | NanoLuc-Y94A luciferase mutant | | Descriptor: | NanoLuc luciferase | | Authors: | Nemergut, M, Marek, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun, 14, 2023
|
|
8AQ6
 
 | |
8AQI
 
 | |
8BO9
 
 | | NanoLuc-D9R/H57A/K89R mutant complexed with azacoelenterazine bound in intra-barrel catalytic site | | Descriptor: | 3-(4-hydroxyphenyl)-8-[(4-hydroxyphenyl)methyl]-5-(phenylmethyl)-1$l^{4},4,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, Non structural polyprotein | | Authors: | Marek, M, Janin, L.Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun, 14, 2023
|
|
8CLT
 
 | |
8CLU
 
 | | Zearalenone lactonase from Rhodococcus erythropolis in complex with zearalactamenone | | Descriptor: | (4~{S})-4-methyl-16,18-bis(oxidanyl)-3-azabicyclo[12.4.0]octadeca-1(18),12,14,16-tetraene-2,8-dione, GLYCEROL, Zearalenone lactonase | | Authors: | Puehringer, D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
