5MQV
 
 | | Crystal structure of human Casein Kinase I delta in complex with 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide | | Descriptor: | 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
6EI1
 
 | | Crystal structure of the covalent complex between deubiquitinase ZUFSP (ZUP1) and Ubiquitin-PA | | Descriptor: | GLYCEROL, MALONATE ION, Polyubiquitin-B, ... | | Authors: | Pichlo, C, Baumann, U, Hofmann, K, Hermanns, T. | | Deposit date: | 2017-09-16 | | Release date: | 2018-03-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A family of unconventional deubiquitinases with modular chain specificity determinants.
Nat Commun, 9, 2018
|
|
5OKT
 
 | | Crystal structure of human Casein Kinase I delta in complex with IWP-2 | | Descriptor: | ACETATE ION, Casein kinase I isoform delta, GLYCEROL, ... | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2017-07-25 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of Inhibitor of Wnt Production 2 (IWP-2) and Related Compounds As Selective ATP-Competitive Inhibitors of Casein Kinase 1 (CK1) delta / epsilon.
J. Med. Chem., 61, 2018
|
|
6HMR
 
 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azothiazole-based inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, MALONIC ACID | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
6HMP
 
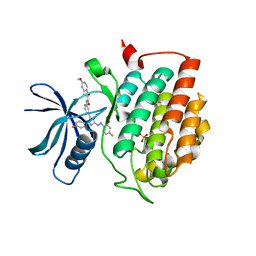 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azoimidazole-based Inhibitor (compound 3) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[5-(4-fluorophenyl)-2-[(~{E})-(4-fluorophenyl)diazenyl]-3-methyl-imidazol-4-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
5LNB
 
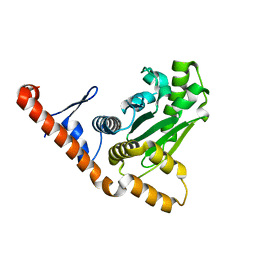 | |
5A0R
 
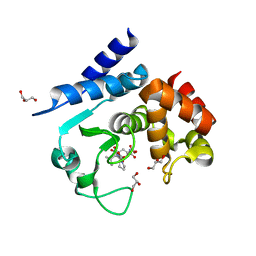 | | Product peptide-bound structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | GLYCEROL, PRODUCT PEPTIDE, ZINC ION, ... | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0P
 
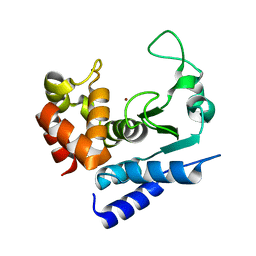 | | Apo-structure of metalloprotease Zmp1 from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0X
 
 | | Substrate peptide-bound structure of metalloprotease Zmp1 variant E143AY178F from Clostridium difficile | | Descriptor: | SUBSTRATE PEPTIDE, ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0S
 
 | | Apo-structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
6SVQ
 
 | | Crystal structure of human GFAT-1 G461E after UDP-GlcNAc soaking | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate-aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6SVM
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate, L-Glu, and UDP-GalNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6SVO
 
 | | Crystal structure of human GFAT-1 in complex with Glucosamine-6-Phosphate and L-Glu | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6SVP
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate, L-Glu, and UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4J
 
 | | Crystal structure of human GFAT-1 G451E in complex with UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4E
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate and L-Glu | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4I
 
 | | Crystal structure of human GFAT-1 G461E | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4F
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4G
 
 | | Crystal structure of human GFAT-1 in complex with UDP-GlcNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, MAGNESIUM ION, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R4H
 
 | | Crystal structure of human GFAT-1 G451E | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-03-22 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6R9Z
 
 | |
8ADD
 
 | | Viral tegument-like DUBs | | Descriptor: | ATP-dependent DNA helicase | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
5ML5
 
 | | Human p38alpha MAPK in complex with imidazolyl pyridine inhibitor 11b | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-methylsulfanyl-1~{H}-imidazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2016-12-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
6HWU
 
 | | Crystal structure of p38alpha in complex with a photoswitchable 2-Azothiazol-based Inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
6HWT
 
 | | Crystal structure of p38alpha in complex with a reduced photoswitchable 2-Azothiazol-based Inhibitor (compound 31) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-(2-phenylhydrazinyl)-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
