3NFM
 
 | | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the complex of type I ribosome inactivating protein with fructose at 2.5A resolution
To be Published
|
|
3RL9
 
 | | Crystal Structure of the complex of type I RIP with the hydrolyzed product of dATP, adenine at 1.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
4HOA
 
 | | Crystal structure of the complex of type 1 ribosome inactivating protein from Momordica Balsamina with B-D-galactopyranosyl-(1-4)-D-glucose at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of type 1 ribosome inactivating protein from Momordica Balsamina with B-D-galactopyranosyl-(1-4)-D-glucose at 2.0 A resolution
To be Published
|
|
3V2K
 
 | | Crystal structure of ribosome inactivating protein from momordica balsamina complexed with the product of RNA substrate adenosine triphosphate at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states.
Biochim.Biophys.Acta, 1824, 2012
|
|
2RGP
 
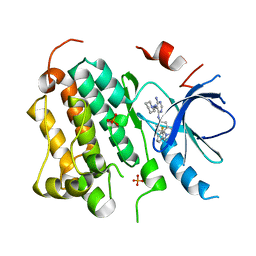 | | Structure of EGFR in complex with hydrazone, a potent dual inhibitor | | Descriptor: | Epidermal growth factor receptor, N-[1-(3-fluorobenzyl)-1H-indazol-5-yl]-5-[(piperidin-1-ylamino)methyl]pyrimidine-4,6-diamine, PHOSPHATE ION | | Authors: | Abad, M.C. | | Deposit date: | 2007-10-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4-Amino-6-arylamino-pyrimidine-5-carbaldehyde hydrazones as potent ErbB-2/EGFR dual kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BEL
 
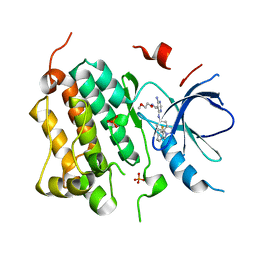 | | X-ray structure of EGFR in complex with oxime inhibitor | | Descriptor: | 4-amino-6-{[1-(3-fluorobenzyl)-1H-indazol-5-yl]amino}pyrimidine-5-carbaldehyde O-(2-methoxyethyl)oxime, Epidermal growth factor receptor, PHOSPHATE ION | | Authors: | Abad, M.C, Xu, G, Neeper, M.P, Struble, G.T, Gaul, M.D, Connolly, P.J. | | Deposit date: | 2007-11-19 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel 4-amino-6-arylaminopyrimidine-5-carbaldehyde oximes as dual inhibitors of EGFR and ErbB-2 protein tyrosine kinases.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3GC1
 
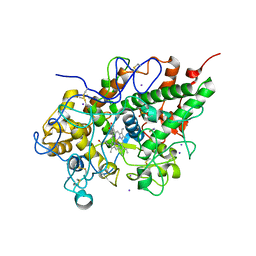 | | Crystal structure of bovine lactoperoxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-21 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 A resolution.
J.Biol.Chem., 285, 2010
|
|
7WYJ
 
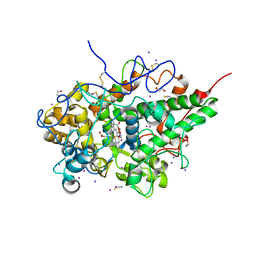 | | Structure of the complex of lactoperoxidase with nitric oxide catalytic product nitrite at 1.89 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Pandey, N, Singh, A.K, Sinha, M, Singh, R.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural evidence of the conversion of nitric oxide (NO) to nitrite ion (NO2-) by lactoperoxidase (LPO): Structure of the complex of LPO with NO2- at 1.89 angstrom resolution
J.Inorg.Biochem., 247, 2023
|
|
3N1N
 
 | | Crystal structure of the complex of type I ribosome inactivating protein with guanine at 2.2A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANINE, Ribosome inactivating protein | | Authors: | Kushwaha, G.S, Singh, N, Sinha, M, Kaur, P, Betzel, C, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-16 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
4FNN
 
 | | Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION | | Descriptor: | Peptidoglycan recognition protein 1, STEARIC ACID | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site.
Arch.Biochem.Biophys., 529, 2013
|
|
3T2V
 
 | | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with mycolic acid at 2.5 A resolution | | Descriptor: | (2S,3R)-2-hexyl-3-hydroxynonanoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3UIL
 
 | | Crystal Structure of the complex of PGRP-S with lauric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, LAURIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3USX
 
 | | Crystal structure of PGRP-S complexed with Myristic Acid at 2.28 A resolution | | Descriptor: | GLYCEROL, MYRISTIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Yamini, S, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-24 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
6L9T
 
 | | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Viswanathan, V, Pandey, N, Singh, A, Sinha, M, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution
To Be Published
|
|
3TGY
 
 | | Crystal structure of the complex of Bovine Lactoperoxidase with Ascorbic acid at 2.35 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Singh, R.P, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution.
Biochim.Biophys.Acta, 1865, 2017
|
|
3V6Q
 
 | | Crystal structure of the complex of bovine lactoperoxidase with Carbon monoxide at 2.0 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bovine carbonyl lactoperoxidase structure at 2.0 angstrom resolution and infrared spectra as a function of pH.
Protein J., 31, 2012
|
|
