7YJK
 
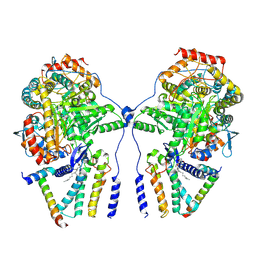 | | Cryo-EM structure of the dimeric atSPT-ORM1 complex | | Descriptor: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJO
 
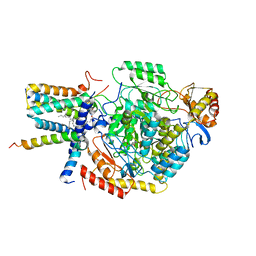 | | Cryo-EM structure of the monomeric atSPT-ORM1 (LCB2a-deltaN5) complex | | Descriptor: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJM
 
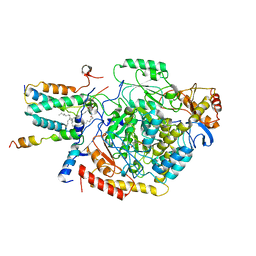 | | Cryo-EM structure of the monomeric atSPT-ORM1 complex | | Descriptor: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJN
 
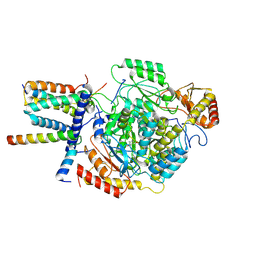 | | Cryo-EM structure of the monomeric atSPT-ORM1 (ORM1-N17A) complex | | Descriptor: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, ORMDL family protein, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-20 | | Release date: | 2023-04-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
8OXG
 
 | | Crystal structure of human methionine aminopeptidase-2 complexed with (3R,4S,5S,6R)-5-methoxy-4-[(2R,3R)-2-methyl-3-(3-methyl-2-buten-1-yl)-2-oxiranyl]-1-oxaspiro[2.5]oct-6-yl N-(trans-4-aminocyclohexyl)carbamate | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, [(1~{R},2~{S},3~{S},4~{R})-2-methoxy-4-methyl-3-[(2~{R},3~{S})-2-methyl-3-(3-methylbut-2-enyl)oxiran-2-yl]-4-oxidanyl-cyclohexyl] ~{N}-(4-azanylcyclohexyl)carbamate | | Authors: | Moss, S, Cornelius, P. | | Deposit date: | 2023-05-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Pharmacological Characterization of SDX-7320/Evexomostat: A Novel Methionine Aminopeptidase Type 2 Inhibitor with Anti-tumor and Anti-metastatic Activity.
Mol.Cancer Ther., 23, 2024
|
|
7F3Q
 
 | | SARS-CoV-2 RBD in complex with A5-10 Fab and A34-2 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A34-2 Fab, ... | | Authors: | Dou, Y, Wang, X, Liu, P, Lu, B, Wang, K. | | Deposit date: | 2021-06-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Development of neutralizing antibodies against SARS-CoV-2, using a high-throughput single-B-cell cloning method.
Antib Ther, 6, 2023
|
|
1REO
 
 | | L-amino acid oxidase from Agkistrodon halys pallas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AHPLAAO, CITRIC ACID, ... | | Authors: | Zhang, H, Teng, M, Niu, L, Wang, Y, Wang, Y, Liu, Q, Huang, Q, Hao, Q, Dong, Y, Liu, P. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Purification, partial characterization, crystallization and structural determination of AHP-LAAO, a novel L-amino-acid oxidase with cell apoptosis-inducing activity from Agkistrodon halys pallas venom.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ZZ6
 
 | | Crystal Structure of Apo-HppE | | Descriptor: | Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
5V54
 
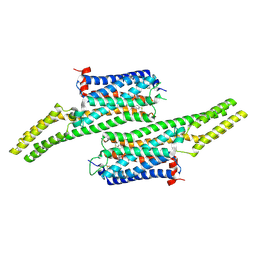 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
1ZZB
 
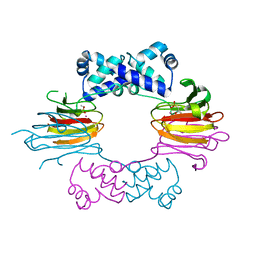 | | Crystal Structure of CoII HppE in Complex with Substrate | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, COBALT (II) ION, Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ8
 
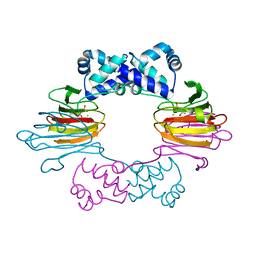 | | Crystal Structure of FeII HppE in Complex with Substrate Form 2 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZC
 
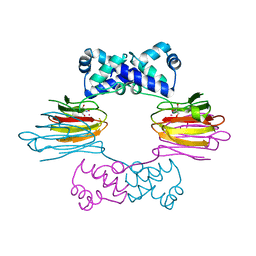 | | Crystal Structure of CoII HppE in Complex with Tris Buffer | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COBALT (II) ION, hydroxypropylphosphonic acid epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ9
 
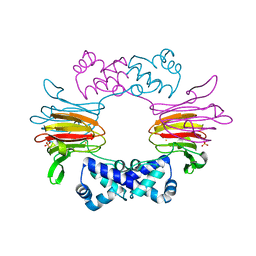 | | Crystal Structure of FeII HppE | | Descriptor: | FE (II) ION, Hydroxypropylphosphonic Acid Epoxidase, SULFATE ION | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
1ZZ7
 
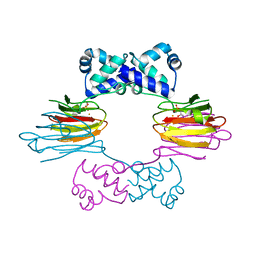 | | Crystal Structure of FeII HppE in Complex with Substrate form 1 | | Descriptor: | (S)-2-HYDROXYPROPYLPHOSPHONIC ACID, FE (II) ION, Hydroxyprophylphosphonic Acid Epoxidase | | Authors: | Higgins, L.J, Yan, F, Liu, P, Liu, H.W, Drennan, C.L. | | Deposit date: | 2005-06-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme
Nature, 437, 2005
|
|
5DO2
 
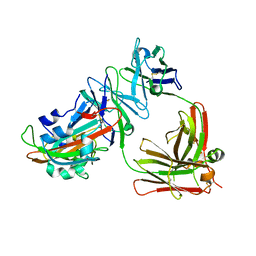 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
7JOA
 
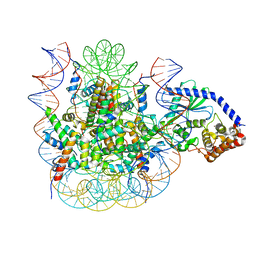 | | 2:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
7JO9
 
 | | 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
8YBN
 
 | | Crystal structure of nanobody SEB-Nb8 bound to staphylococcal enterotoxin B (SEB) | | Descriptor: | Enterotoxin type B, nanobody SEB-Nb8 | | Authors: | Ding, Y, Zong, X, Liu, R, Liu, P. | | Deposit date: | 2024-02-15 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Int.J.Biol.Macromol., 276, 2024
|
|
8YBO
 
 | | Crystal structure of nanobody SEB-Nb11 bound to staphylococcal enterotoxin B (SEB) | | Descriptor: | Enterotoxin type B, nanobody SEB-Nb11 | | Authors: | Ding, Y, Zong, X, Liu, R, Liu, P. | | Deposit date: | 2024-02-15 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Int.J.Biol.Macromol., 276, 2024
|
|
8YBM
 
 | | Crystal structure of nanobody SEB-Nb6 bound to staphylococcal enterotoxin B (SEB) | | Descriptor: | Enterotoxin type B, nanobody SEB-Nb6 | | Authors: | Ding, Y, Zong, X, Liu, R, Liu, P. | | Deposit date: | 2024-02-15 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Int.J.Biol.Macromol., 276, 2024
|
|
7EMD
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-GLY-ASP-PHE-PHE-HIS-ASP-MET-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMC
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-THR-GLU-ILE-ARG-GLU-LEU-LEU-VAL, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMB
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | ALA-ALA-ALA-ILE-GLU-GLU-GLU-ASP-ILE, Beta-2-microglobulin, Leucocyte antigen | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EMA
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, TYR-SER-SER-ASP-VAL-THR-THR-LEU-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
7EM9
 
 | | Mooring Stone-Like Arg114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I | | Descriptor: | Beta-2-microglobulin, Leucocyte antigen, SER-LEU-ASP-GLU-TYR-SER-SER-ASP-VAL | | Authors: | Yue, C, Xiang, W, Huang, X, Sun, Y, Xiao, J, Liu, K, Sun, Z, Qiao, P, Li, H, Gan, J, Ba, L, Chai, Y, Qi, J, Liu, P, Qi, P, Zhao, Y, Li, Y, Qiu, H.J, Gao, G.F, Gao, G, Liu, W.J. | | Deposit date: | 2021-04-13 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mooring Stone-Like Arg 114 Pulls Diverse Bulged Peptides: First Insight into African Swine Fever Virus-Derived T Cell Epitopes Presented by Swine Major Histocompatibility Complex Class I.
J.Virol., 96, 2022
|
|
