7XIN
 
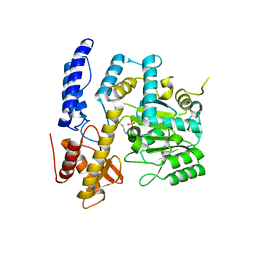 | | Crystal structure of DODC from Pseudomonas | | Descriptor: | DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Li, X, Zhou, Y.L, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Sun, D.Y, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of DODC from Pseudomonas
To Be Published
|
|
1CS3
 
 | |
9IKV
 
 | | human SLC26A7 with iodide | | Descriptor: | Anion exchange transporter, IODIDE ION | | Authors: | Li, X, Yang, X, Lu, X, Lin, B, Zhang, Y, Huang, B, Zhou, Y, Huang, J, Wu, K, Zhou, Q, Chi, X. | | Deposit date: | 2024-06-29 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | human SLC26A7 with iodide
To Be Published
|
|
9IKX
 
 | | human SLC26A7 apo state | | Descriptor: | Anion exchange transporter | | Authors: | Li, X, Yang, X, Lu, X, Lin, B, Zhang, Y, Huang, B, Zhou, Y, Huang, J, Wu, K, Zhou, Q, Chi, X. | | Deposit date: | 2024-06-29 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | human SLC26A7 apo state
To Be Published
|
|
3T7I
 
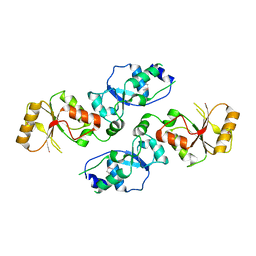 | | Crystal structure of Se-Met Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
3T7K
 
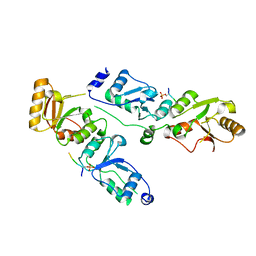 | | Complex structure of Rtt107p and phosphorylated histone H2A | | Descriptor: | Histone H2A.1, Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
3T7J
 
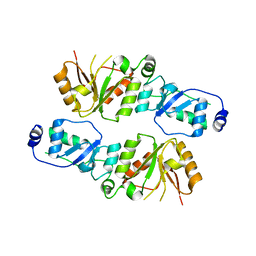 | | Crystal structure of Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
7YMN
 
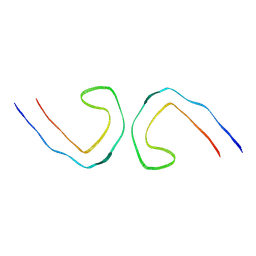 | | Cryo-EM structure of in vitro PHF fibril | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Li, X, Liu, C. | | Deposit date: | 2022-07-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Subtle change of fibrillation condition leads to substantial alteration of recombinant Tau fibril structure.
Iscience, 25, 2022
|
|
9LMX
 
 | | Inactive mutant of FAST-ACC-T140E/R132G in complex with mono(2-hydroxyethyl) terephthalic acid | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, Ning, Z.Y, Huang, S.Q, Zeng, C, Zeng, Z.Y, Ji, R, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2025-01-20 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Combined approaches to enhance the Pichia pastoris-expressed PET hydrolase.
Int.J.Biol.Macromol., 2025
|
|
5H0S
 
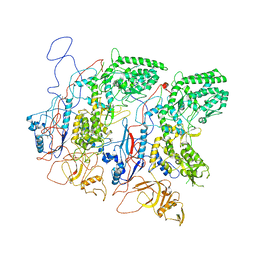 | | EM Structure of VP1A and VP1B | | Descriptor: | VP1 | | Authors: | Li, X, Zhou, N, Xu, B, Chen, W, Zhu, B, Wang, X, Wang, J, Liu, H, Cheng, L. | | Deposit date: | 2016-10-06 | | Release date: | 2017-01-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
5H0R
 
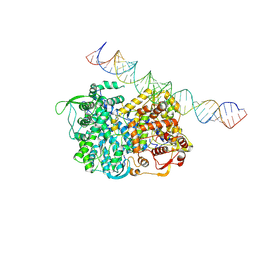 | | RNA dependent RNA polymerase ,vp4,dsRNA | | Descriptor: | RNA (42-MER), RNA-dependent RNA polymerase, VP4 protein | | Authors: | Li, X, Zhou, N, Chen, W, Zhu, B, Wang, X, Xu, B, Wang, J, Liu, H, Cheng, L. | | Deposit date: | 2016-10-06 | | Release date: | 2017-01-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
9LMS
 
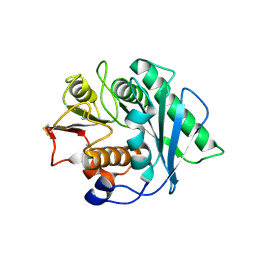 | | Crystal structure of Pichia pastoris-expressed FAST-PETase-N212A/K233C/S282C variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, Ning, Z.Y, Huang, S.Q, Zeng, C, Zeng, Z.Y, Ji, R, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2025-01-20 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Combined approaches to enhance the Pichia pastoris-expressed PET hydrolase.
Int.J.Biol.Macromol., 2025
|
|
9LMU
 
 | | Crystal structure of variant FAST-ACC-A47E | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, Ning, Z.Y, Huang, S.Q, Zeng, C, Zeng, Z.Y, Ji, R, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2025-01-20 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Combined approaches to enhance the Pichia pastoris-expressed PET hydrolase.
Int.J.Biol.Macromol., 2025
|
|
9LMT
 
 | | Crystal structure of variant FAST-ACC-A248E | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, Ning, Z.Y, Huang, S.Q, Zeng, C, Zeng, Z.Y, Ji, R, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2025-01-20 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combined approaches to enhance the Pichia pastoris-expressed PET hydrolase.
Int.J.Biol.Macromol., 2025
|
|
9LMW
 
 | | Crystal structure of variant FAST-ACC-T140E | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, Ning, Z.Y, Huang, S.Q, Zeng, C, Zeng, Z.Y, Ji, R, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2025-01-20 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combined approaches to enhance the Pichia pastoris-expressed PET hydrolase.
Int.J.Biol.Macromol., 2025
|
|
9LMV
 
 | | Crystal structure of FAST-ACC-T140D/R132G/S160A mutant in complex with mono(2-hydroxyethyl) terephthalic acid | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, Ning, Z.Y, Huang, S.Q, Zeng, C, Zeng, Z.Y, Ji, R, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2025-01-20 | | Release date: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Combined approaches to enhance the Pichia pastoris-expressed PET hydrolase.
Int.J.Biol.Macromol., 2025
|
|
9F3D
 
 | |
5U73
 
 | | Crystal structure of human Niemann-Pick C1 protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Niemann-Pick C1 protein, ... | | Authors: | Li, X, Wang, J, Blobel, G. | | Deposit date: | 2016-12-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.348 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U74
 
 | | Structure of human Niemann-Pick C1 protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5X5S
 
 | |
5VB3
 
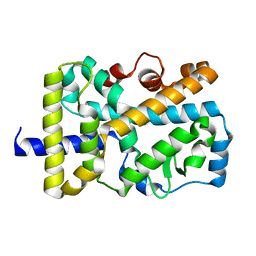 | | X-ray structure of nuclear receptor ROR-gammat Ligand Binding Domain + SRC2 peptide | | Descriptor: | Nuclear receptor ROR-gamma, SRC2 chimera, SODIUM ION | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
5V5M
 
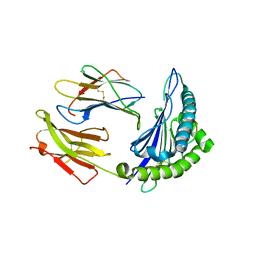 | |
5VB7
 
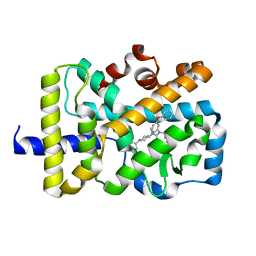 | | X-ray co-structure of nuclear receptor ROR-gammat Ligand Binding Domain with an agonist and SRC2 peptide | | Descriptor: | N-methyl-N'-(3-methylbut-2-en-1-yl)-N'-(3-phenoxyphenyl)-N-[trans-4-(pyridin-4-yl)cyclohexyl]urea, Nuclear receptor ROR-gamma, SRC2 chimera, ... | | Authors: | Li, X. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Structural studies unravel the active conformation of apo ROR gamma t nuclear receptor and a common inverse agonism of two diverse classes of ROR gamma t inhibitors.
J. Biol. Chem., 292, 2017
|
|
1ZVF
 
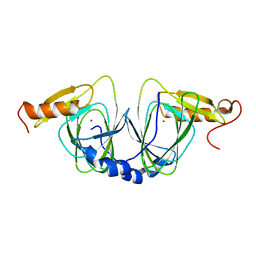 | | The crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from Saccharomyces cerevisiae | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, NICKEL (II) ION | | Authors: | Li, X, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from Saccharomyces cerevisiae
To be published
|
|
2DC2
 
 | | Solution Structure of PDZ Domain | | Descriptor: | golgi associated PDZ and coiled-coil motif containing isoform b | | Authors: | Li, X, Wu, J, Shi, Y. | | Deposit date: | 2005-12-20 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of GOPC PDZ domain and its interaction with the C-terminal motif of neuroligin
Protein Sci., 15, 2006
|
|
